Difference between revisions of "Assembly:RBS-CDS issues"
m |
|||
| (13 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | [[Category:BioBrick RFC 10]] | |
| + | {{HelpPage/MainLinks}} | ||
| + | == Transfer Curves == | ||
| − | The transfer curve of a BioBrick device is measured from input PoPs to output PoPs. For a protein-DNA inhibition system, | + | The [http://openwetware.org/wiki/Endy:F2620/Transfer_Curve transfer curve] of a BioBrick device is measured from input PoPs to output PoPs. (More on PoPs [http://parts2.mit.edu/wiki/index.php/Abstraction_hierarchy_and_PoPS here].) For a protein-DNA inhibition system, the transfer curve depends on the rates and efficiencies of many stages in a chain that includes: |
| + | |||
| + | *transcriptional efficiency | ||
| + | *mRNA lifetime | ||
| + | *ribosome binding efficiency | ||
| + | *translation initiation and efficiency | ||
| + | *protein lifetime | ||
| + | *protein concentration | ||
| + | *protein multimerization | ||
| + | *protein-DNA binding rates and efficiencies | ||
| + | *cooperative effects with other molecules | ||
| + | *RNA polymerase effects | ||
| + | *system copy count | ||
| + | |||
| + | and more system-specific variables, few if any of which can be measured directly. | ||
If BioBrick parts are to be interoperable, intentional engineering of this chain of events will be required to produce a desired transfer function. | If BioBrick parts are to be interoperable, intentional engineering of this chain of events will be required to produce a desired transfer function. | ||
| − | + | == Ribosome Binding Sites (RBS) == | |
| + | |||
| + | Different RBS bind ribosomes with different efficiencies. While the overall efficiency may not be directly proportional to this efficiency, it is an important variable. In the future, we expect that this data book will have a table such as the following: | ||
'''Standard BioBrick Ribosome Binding Sites''' | '''Standard BioBrick Ribosome Binding Sites''' | ||
| Line 22: | Line 40: | ||
| − | A BioBrick engineer armed with this table could perform a first-order reengineering of the transfer function by measuring a component with BBa_B0031 and then selecting a more appropriate RBS. | + | A BioBrick engineer armed with this table could perform a first-order reengineering of the transfer function by measuring a component with [https://parts.igem.org/wiki/index.php/Part:BBa_B0031 BBa_B0031] and then selecting a more appropriate RBS. |
| − | In order to allow easy variation in the RBS, some BioBrick | + | In order to allow easy variation in the RBS, some BioBrick parts have their RBS and their protein coding regions implemented in separate parts. The combined, normalized part is also available for convenience. In order to divide the BioBrick part into its RBS and coding region, a set of BioBrick restriction sites must be designed. |
== Standard Assembly and the RBS == | == Standard Assembly and the RBS == | ||
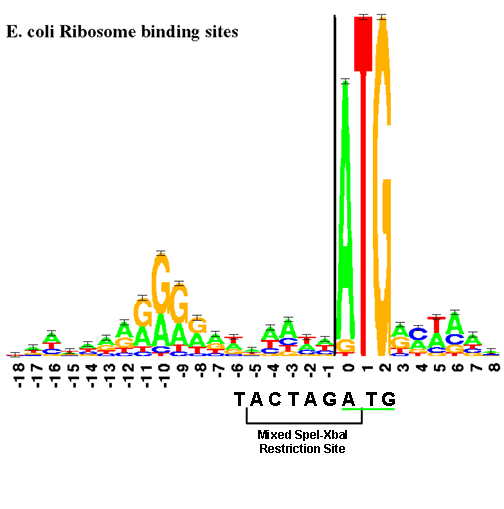
| − | The RBS is physically near the start codon ( | + | The RBS is physically near the start codon. (In BioBricks, this will always be ATG.) The "sequence logo", similar to a consensus sequence, for the RBS is shown below. The size of each letter is proportional to the frequency of that base in the RBS. |
| − | + | ||
| + | [[Image:Ribologo-small.gif|frame|center|The RBS sequence logo, from Tom Schneider, [http://www-lmmb.ncifcrf.gov/%7Etoms/sequencelogo.html "A Gallery of Sequence Logos"]]] | ||
| + | |||
| + | This figure shows the RBS as an A/G-rich region about 10 bases upstream of the start codon. This leaves little room for a BioBrick restriction enzyme site. The standard assembly method of BioBricks normally results in an 8-base region containing a 6-base mixed SpeI/XbaI restriction site surrounded by a base on each end as shown below. If a start codon followed this sequence, the RBS would be significantly disturbed. | ||
| + | |||
| + | |||
| + | |||
| + | --T ACTAGA G-- | ||
| − | |||
| − | |||
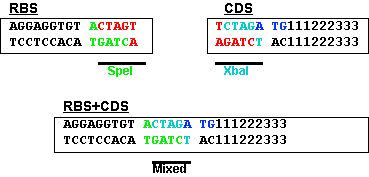
This problem is solved by changing the rightmost base from a G to a T. This results in the sequence shown below and limits the impact of the BioBrick overhead to six bases. | This problem is solved by changing the rightmost base from a G to a T. This results in the sequence shown below and limits the impact of the BioBrick overhead to six bases. | ||
| − | Summary | + | |
| + | [[image:RBS-CDS.gif|frame|center]] | ||
| + | |||
| + | == Summary == | ||
When variable RBS sequences are developed as BioBrick parts, they should have the standard BioBrick prefix and suffix as follows: | When variable RBS sequences are developed as BioBrick parts, they should have the standard BioBrick prefix and suffix as follows: | ||
| − | |||
| − | |||
| − | |||
| − | Variable RBS Suffix | + | {| border="1" cellpadding="0" |
| + | |- | ||
| + | !width="150"|Variable RBS Prefix | ||
| + | | | ||
| + | |||
| + | GAATTC GCGGCCGC T TCTAGA G[RBS] | ||
| + | EcoRI NotI XbaI | ||
| + | |||
| + | |- | ||
| + | !width="150"|Variable RBS Suffix | ||
| + | | | ||
| + | |||
| + | [RBS]T ACTAGT A GCGGCCG CTGCAG | ||
| + | SpeI NotI PstI | ||
| + | |||
| + | |- | ||
| + | |} | ||
| − | |||
| − | |||
When a protein coding region is designed to be preceded by a BioBrick RBS part, it should have the following prefix and suffix sequences: | When a protein coding region is designed to be preceded by a BioBrick RBS part, it should have the following prefix and suffix sequences: | ||
| − | |||
| − | |||
| − | |||
| − | Protein Coding Region | + | {| border="1" cellpadding="0" |
| + | |- | ||
| + | !width="150"|Protein Coding Region Prefix* | ||
| + | | | ||
| − | [CDS] | + | GAATTC GCGGCCGC T TCTAG[ATG Remaining CDS] |
| − | + | EcoRI NotI XbaI | |
| − | Notes | + | |- |
| + | !width="150"|Protein Coding Region Suffix | ||
| + | | | ||
| + | |||
| + | [CDS]T ACTAGT A GCGGCCG CTGCAG | ||
| + | SpeI NotI PstI | ||
| + | |||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | == Notes == | ||
*The only change is that the protein coding region prefix does NOT end with AGAG. instead, it ends AGATG with the ATG also being the start codon of the protein coding region. | *The only change is that the protein coding region prefix does NOT end with AGAG. instead, it ends AGATG with the ATG also being the start codon of the protein coding region. | ||
| − | In BioBrick parts, we prefer using TAA as the stop codon. | + | *In BioBrick parts, we prefer using TAA as the stop codon. |
| + | |||
| + | |||
| + | <hr> | ||
| + | <small> | ||
| + | |||
| + | *Back to [[Assembly|assembly]] | ||
| + | |||
| + | See also: | ||
| + | |||
| + | *[[Assembly:Parts in plasmids|Parts in Plasmids]] | ||
| + | |||
| + | *[[Assembly:Restriction enzymes|Restriction Enzymes]] | ||
| + | |||
| + | *[[Assembly:Rolling assembly|Rolling Assembly]] | ||
| + | |||
| + | </small> | ||
Latest revision as of 19:51, 16 June 2017
- Registry Help Pages:
- TOC
- At-a-Glance
- FAQ
Contents
Transfer Curves
The [http://openwetware.org/wiki/Endy:F2620/Transfer_Curve transfer curve] of a BioBrick device is measured from input PoPs to output PoPs. (More on PoPs [http://parts2.mit.edu/wiki/index.php/Abstraction_hierarchy_and_PoPS here].) For a protein-DNA inhibition system, the transfer curve depends on the rates and efficiencies of many stages in a chain that includes:
- transcriptional efficiency
- mRNA lifetime
- ribosome binding efficiency
- translation initiation and efficiency
- protein lifetime
- protein concentration
- protein multimerization
- protein-DNA binding rates and efficiencies
- cooperative effects with other molecules
- RNA polymerase effects
- system copy count
and more system-specific variables, few if any of which can be measured directly.
If BioBrick parts are to be interoperable, intentional engineering of this chain of events will be required to produce a desired transfer function.
Ribosome Binding Sites (RBS)
Different RBS bind ribosomes with different efficiencies. While the overall efficiency may not be directly proportional to this efficiency, it is an important variable. In the future, we expect that this data book will have a table such as the following:
Standard BioBrick Ribosome Binding Sites
| Part Number | Binding Efficiency | Sequence |
|---|---|---|
| BBa_B0030 | 0.6 | att aaa gag gag aaa |
| BBa_B0031 | 0.07 | tca cac agg aaa cc |
| BBa_B0032 | 0.3 | tca cac agg aaa g |
A BioBrick engineer armed with this table could perform a first-order reengineering of the transfer function by measuring a component with BBa_B0031 and then selecting a more appropriate RBS.
In order to allow easy variation in the RBS, some BioBrick parts have their RBS and their protein coding regions implemented in separate parts. The combined, normalized part is also available for convenience. In order to divide the BioBrick part into its RBS and coding region, a set of BioBrick restriction sites must be designed.
Standard Assembly and the RBS
The RBS is physically near the start codon. (In BioBricks, this will always be ATG.) The "sequence logo", similar to a consensus sequence, for the RBS is shown below. The size of each letter is proportional to the frequency of that base in the RBS.
This figure shows the RBS as an A/G-rich region about 10 bases upstream of the start codon. This leaves little room for a BioBrick restriction enzyme site. The standard assembly method of BioBricks normally results in an 8-base region containing a 6-base mixed SpeI/XbaI restriction site surrounded by a base on each end as shown below. If a start codon followed this sequence, the RBS would be significantly disturbed.
--T ACTAGA G--
This problem is solved by changing the rightmost base from a G to a T. This results in the sequence shown below and limits the impact of the BioBrick overhead to six bases.
Summary
When variable RBS sequences are developed as BioBrick parts, they should have the standard BioBrick prefix and suffix as follows:
| Variable RBS Prefix |
GAATTC GCGGCCGC T TCTAGA G[RBS]
EcoRI NotI XbaI
|
|---|---|
| Variable RBS Suffix |
[RBS]T ACTAGT A GCGGCCG CTGCAG
SpeI NotI PstI
|
When a protein coding region is designed to be preceded by a BioBrick RBS part, it should have the following prefix and suffix sequences:
| Protein Coding Region Prefix* |
GAATTC GCGGCCGC T TCTAG[ATG Remaining CDS]
EcoRI NotI XbaI
|
|---|---|
| Protein Coding Region Suffix |
[CDS]T ACTAGT A GCGGCCG CTGCAG
SpeI NotI PstI
|
Notes
- The only change is that the protein coding region prefix does NOT end with AGAG. instead, it ends AGATG with the ATG also being the start codon of the protein coding region.
- In BioBrick parts, we prefer using TAA as the stop codon.
- Back to assembly
See also: