Difference between revisions of "Help:Restriction enzymes"
| (10 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Enzymes.png|left]] | [[Image:Enzymes.png|left]] | ||
| − | Microbiologists and molecular biologists regularly use restriction enzymes to cut and splice DNA. ''Note:'' To learn more about restriction enzymes, see the [http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/default.asp New England Bioloabs website]. | + | Microbiologists and molecular biologists regularly use restriction enzymes to cut and splice DNA. <small>''Note:'' To learn more about restriction enzymes, see the [http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/default.asp New England Bioloabs website].</small> |
| − | |||
| − | + | Restriction enzymes work by recognizing a particular sequence of bases on the DNA. The enzyme then cuts the backbones of both strands, allowing the DNA to separate into two pieces. | |
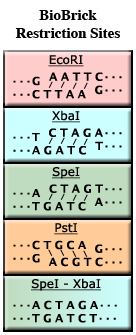
| − | + | For example, the enzyme EcoRI (see the figure, left, top) binds to the recognition sequence '''GAATTC''' and cuts between the G and the A. It also cuts between the A and the G on the complementary strand. Because EcoRI does not cut across both strands of DNA in the same place, four unmatched bases of DNA will ''stick out'' of each end (AATT in our example). These four bases of single-stranded DNA are said to be ''sticky'' because they have the ability to stick to a matching piece of single-stranded DNA. | |
| + | Thus, if you want to move a piece of DNA from one plasmid to another, you can use restriction enzymes to cut it out of one plasmid, then use the same restriction enzymes to create a gap in the destination plasmid, purify the desired segments, and mix the results. The sticky ends will "anneal" together via base pairing and a ligase enzyme can be used to reseal the backbones. After ligation, if everything has worked properly, the DNA pieces will be joined together. If two pieces of DNA are cut by the same enzyme, ligation will re-form the original restriction site, and if the restriction enzyme is used again, the pieces will be cut apart in the same way. | ||
| − | '''SpeI - XbaI Mixed Sites:'''<br> | + | '''SpeI - XbaI Mixed Sites and BioBrick Assembly:'''<br> |
| − | However, with [[Assembly: | + | However, with [[Assembly:BioBrick_standard_assembly|BioBrick Standard Assembly]], we want to be able to assemble two parts and then use the resulting assembly as a new part in the same process. To do this, we use two enzymes that have ''compatible'' sticky ends but ''incompatible'' recognition sequences, like SpeI and XbaI. |
| − | Note that both XbaI and SpeI have the same sticky ends, '''CTAG'''. As a result, DNA cut by one enzyme can stick to DNA cut by the other enzyme. However, after ligation, the resulting sequence does not match the recognition site of either XbaI or SpeI (see bottom of figure). This is called a '''mixed site'''. The formation of mixed sites is the basis of [[Assembly: | + | Note that both XbaI and SpeI have the same sticky ends, '''CTAG'''. As a result, DNA cut by one enzyme can stick to DNA cut by the other enzyme. However, after ligation, the resulting sequence does not match the recognition site of either XbaI or SpeI (see bottom of figure). This is called a '''mixed site'''. The formation of mixed sites is the basis of [[Assembly:BioBrick_standard_assembly|BioBrick Standard Assembly]]. |
With BioBrick Standard Assembly, four restriction enzymes (XbaI, SpeI, EcoRI, PstI) are sufficient to perform all DNA manipulations. Of course, because these four restriction enzymes are used in the assembly process, BioBrick parts must not contain any of these four restriction sites. The Registry tools check the DNA sequences of all new BioBrick parts to enforce this requirement. | With BioBrick Standard Assembly, four restriction enzymes (XbaI, SpeI, EcoRI, PstI) are sufficient to perform all DNA manipulations. Of course, because these four restriction enzymes are used in the assembly process, BioBrick parts must not contain any of these four restriction sites. The Registry tools check the DNA sequences of all new BioBrick parts to enforce this requirement. | ||
| + | |||
| + | <hr> | ||
| + | <small> | ||
| + | |||
| + | *Back to [[Assembly|assembly]] | ||
| + | |||
| + | See also: | ||
| + | |||
| + | *[[Assembly:Parts in plasmids|Parts in Plasmids]] | ||
| + | |||
| + | *[[Assembly:Rolling assembly|Rolling Assembly]] | ||
| + | |||
| + | *[[Assembly:RBS-CDS issues|RBS-CDS Design Issues]] | ||
| + | |||
| + | </small> | ||
Latest revision as of 18:43, 28 June 2017
Microbiologists and molecular biologists regularly use restriction enzymes to cut and splice DNA. Note: To learn more about restriction enzymes, see the [http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/default.asp New England Bioloabs website].
Restriction enzymes work by recognizing a particular sequence of bases on the DNA. The enzyme then cuts the backbones of both strands, allowing the DNA to separate into two pieces.
For example, the enzyme EcoRI (see the figure, left, top) binds to the recognition sequence GAATTC and cuts between the G and the A. It also cuts between the A and the G on the complementary strand. Because EcoRI does not cut across both strands of DNA in the same place, four unmatched bases of DNA will stick out of each end (AATT in our example). These four bases of single-stranded DNA are said to be sticky because they have the ability to stick to a matching piece of single-stranded DNA.
Thus, if you want to move a piece of DNA from one plasmid to another, you can use restriction enzymes to cut it out of one plasmid, then use the same restriction enzymes to create a gap in the destination plasmid, purify the desired segments, and mix the results. The sticky ends will "anneal" together via base pairing and a ligase enzyme can be used to reseal the backbones. After ligation, if everything has worked properly, the DNA pieces will be joined together. If two pieces of DNA are cut by the same enzyme, ligation will re-form the original restriction site, and if the restriction enzyme is used again, the pieces will be cut apart in the same way.
SpeI - XbaI Mixed Sites and BioBrick Assembly:
However, with BioBrick Standard Assembly, we want to be able to assemble two parts and then use the resulting assembly as a new part in the same process. To do this, we use two enzymes that have compatible sticky ends but incompatible recognition sequences, like SpeI and XbaI.
Note that both XbaI and SpeI have the same sticky ends, CTAG. As a result, DNA cut by one enzyme can stick to DNA cut by the other enzyme. However, after ligation, the resulting sequence does not match the recognition site of either XbaI or SpeI (see bottom of figure). This is called a mixed site. The formation of mixed sites is the basis of BioBrick Standard Assembly.
With BioBrick Standard Assembly, four restriction enzymes (XbaI, SpeI, EcoRI, PstI) are sufficient to perform all DNA manipulations. Of course, because these four restriction enzymes are used in the assembly process, BioBrick parts must not contain any of these four restriction sites. The Registry tools check the DNA sequences of all new BioBrick parts to enforce this requirement.
- Back to assembly
See also: