Difference between revisions of "Part:BBa K771001"
(→Design) |
|||
| (35 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K771001 short</partinfo> | <partinfo>BBa_K771001 short</partinfo> | ||
| − | RNA aptamer binding protein PP7 fused with membrane protein Lgt | + | Membrane anchor 1:RNA aptamer binding protein PP7 ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771111 BBa_K771111]) fused with membrane protein Lgt([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771102 BBa_K771102]). |
| + | |||
| + | ==Overview: Membrane Scaffold System== | ||
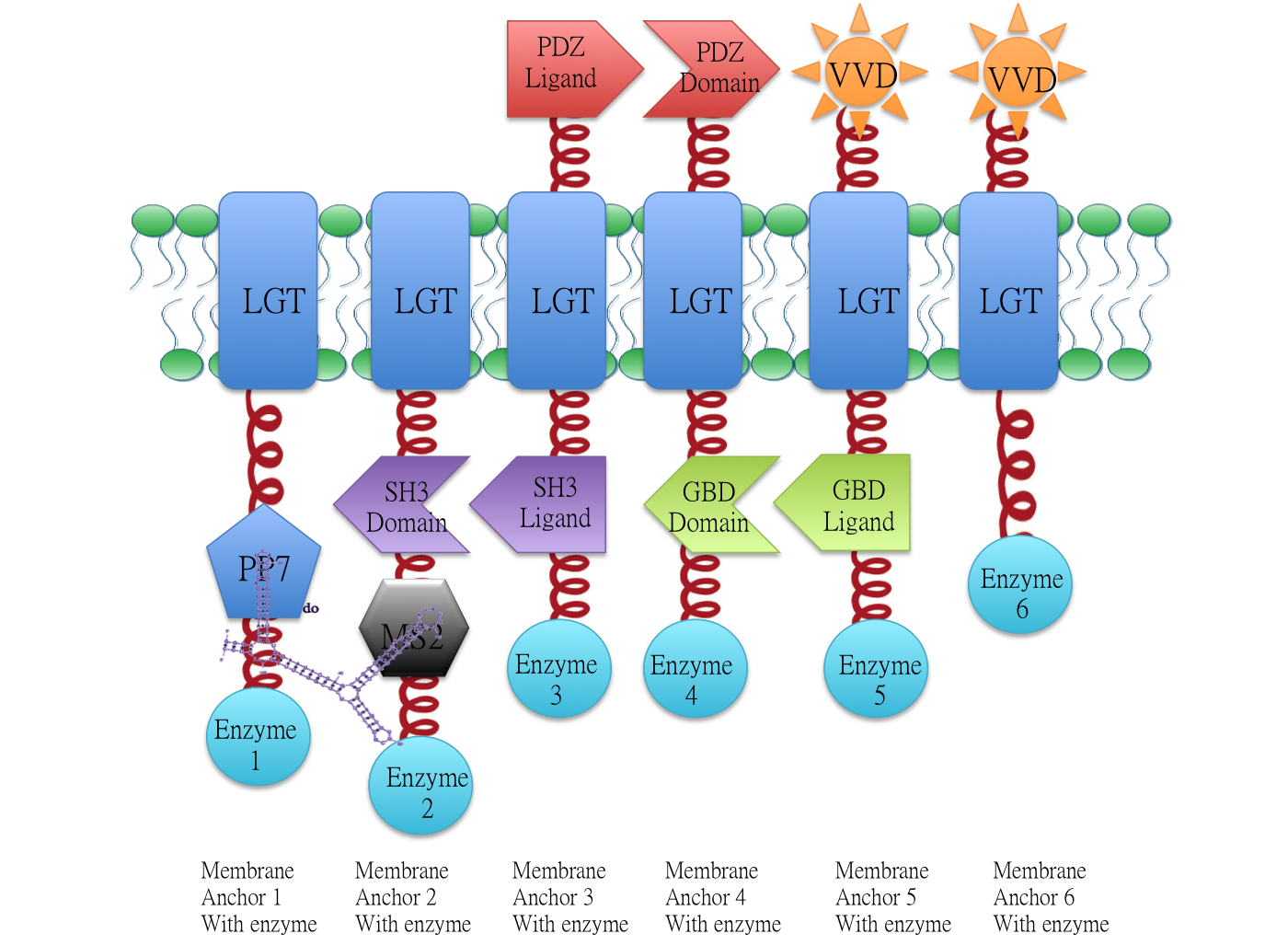
[[Image:12SJTU-MA1-MA6.png|500px|center]] | [[Image:12SJTU-MA1-MA6.png|500px|center]] | ||
| + | Demonstration of the whole ''Membrane Scaffold'' device. Membrane Anchor 2, 3, 4, 5 consitutively aggregate while Membrane Anchor 1 and 6 only aggregate with constitutive cluster (Membrane Anchor 2, 3, 4, 5) when certain signal is present. | ||
| + | ==Backgroud== | ||
| − | + | Membrane Anchor 1 and 2 act as a controlling point to determine whether certain pathway (in this case involving enzyme 1, 2, 3, 4 and 5) is activated or not. RNA molecule D0([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771009 BBa_K771009]) with PP7 and MS2 aptamer domain acts as the signal. | |
| − | + | ||
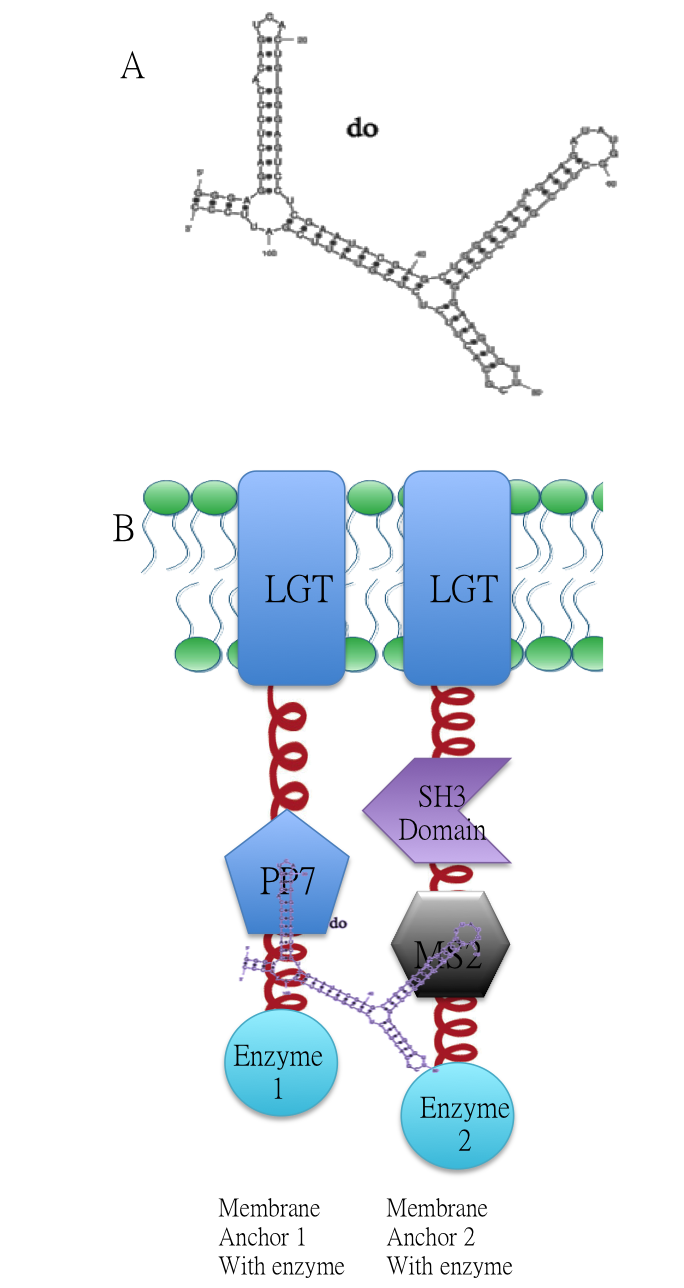
| − | Related Biobrick | + | [[Image:12SJTU RNASIGNAL.png|200px|center]] |
| + | <br/>A:Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. B: Through fusing PP7 and MS2 RNA biding domain ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771111 BBa_K771111] [https://parts.igem.org/wiki/index.php?title=Part:BBa_K771112 BBa_K771112]) to our membrane device, RNA molecule D0([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771009 BBa_K771009]) in Figure A can function as a bridge to connect different proteins, thus decreasing distance between corresponding enzymes. | ||
| + | |||
| + | ==Design== | ||
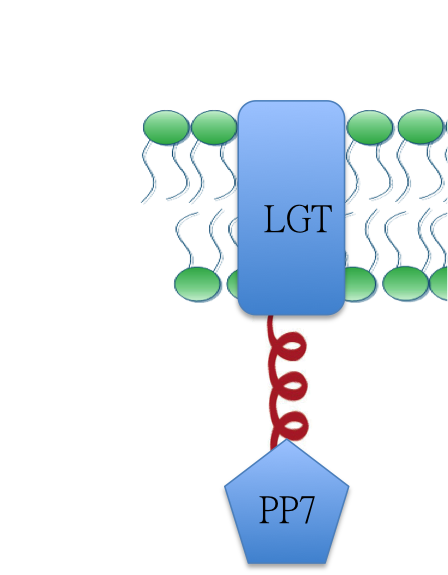
| + | [[Image:12SJTU_MA1.png|200px|center]] | ||
| + | RNA aptamer binding protein PP7([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771111 BBa_K771111]) fused with membrane protein Lgt ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771102 BBa_K771102]). When RNA molecule D0 is present, Membrane Anchor 1 is expected to dimerize with Membrane Anchor 2([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771002 BBa_K771002]) through MS2([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771112 BBa_K771112]) and PP7([https://parts.igem.org/wiki/index.php?title=Part:BBa_K771111 BBa_K771111]) domain. | ||
| + | |||
| + | ==Related Biobrick== | ||
<br\>Membrabe Anchor 2([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771002 BBa_K771002]]) | <br\>Membrabe Anchor 2([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771002 BBa_K771002]]) | ||
<br\>Membrabe Anchor 3([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771003 BBa_K771003]]) | <br\>Membrabe Anchor 3([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771003 BBa_K771003]]) | ||
| Line 15: | Line 25: | ||
<br\>Membrabe Anchor 5([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771005 BBa_K771005]]) | <br\>Membrabe Anchor 5([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771005 BBa_K771005]]) | ||
<br\>Membrabe Anchor 6([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771006 BBa_K771006]]) | <br\>Membrabe Anchor 6([[https://parts.igem.org/wiki/index.php?title=Part:BBa_K771006 BBa_K771006]]) | ||
| + | |||
| + | |||
| + | ==Reference== | ||
| + | Delebecque, C. J., A. B. Lindner, et al. (2011). "Organization of intracellular reactions with rationally designed RNA assemblies." Science 333(6041): 470. | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| − | + | ==Usage and Biology== | |
<!-- --> | <!-- --> | ||
Latest revision as of 01:12, 30 September 2012
Membrane Anchor 1 :ssDsbA-Lgt-PP7
Membrane anchor 1:RNA aptamer binding protein PP7 (BBa_K771111) fused with membrane protein Lgt(BBa_K771102).
Overview: Membrane Scaffold System
Demonstration of the whole Membrane Scaffold device. Membrane Anchor 2, 3, 4, 5 consitutively aggregate while Membrane Anchor 1 and 6 only aggregate with constitutive cluster (Membrane Anchor 2, 3, 4, 5) when certain signal is present.
Backgroud
Membrane Anchor 1 and 2 act as a controlling point to determine whether certain pathway (in this case involving enzyme 1, 2, 3, 4 and 5) is activated or not. RNA molecule D0(BBa_K771009) with PP7 and MS2 aptamer domain acts as the signal.
A:Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. B: Through fusing PP7 and MS2 RNA biding domain (BBa_K771111 BBa_K771112) to our membrane device, RNA molecule D0(BBa_K771009) in Figure A can function as a bridge to connect different proteins, thus decreasing distance between corresponding enzymes.
Design
RNA aptamer binding protein PP7(BBa_K771111) fused with membrane protein Lgt (BBa_K771102). When RNA molecule D0 is present, Membrane Anchor 1 is expected to dimerize with Membrane Anchor 2(BBa_K771002) through MS2(BBa_K771112) and PP7(BBa_K771111) domain.
Related Biobrick
<br\>Membrabe Anchor 2([BBa_K771002]) <br\>Membrabe Anchor 3([BBa_K771003]) <br\>Membrabe Anchor 4([BBa_K771004]) <br\>Membrabe Anchor 5([BBa_K771005]) <br\>Membrabe Anchor 6([BBa_K771006])
Reference
Delebecque, C. J., A. B. Lindner, et al. (2011). "Organization of intracellular reactions with rationally designed RNA assemblies." Science 333(6041): 470.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1129
Illegal BglII site found at 1385 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 883
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 504