Difference between revisions of "Part:BBa K801077"
Nadine1990 (Talk | contribs) (→LC/MS) |
|||
| (10 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K801077 short</partinfo> | <partinfo>BBa_K801077 short</partinfo> | ||
| − | This BioBrick is the final "caffeine synthesis device". It contains all three necessary enzymes: CaXMT1, CaMXMT1 and CaDXMT1, i.e. it is made up of the three single expression cassettes for each enzyme (see also [https://parts.igem.org/Part:BBa_K801073 BBa_K801073], [https://parts.igem.org/Part:BBa_K801074 BBa_K801074 and [https://parts.igem.org/Part:BBa_K801075 BBa_K801075]). It can be directly transformed into competent yeast cells or cloned in an adequate yeast genome integration vector. | + | This BioBrick is the final "caffeine synthesis device". It contains all three necessary enzymes: CaXMT1, CaMXMT1 and CaDXMT1, i.e. it is made up of the three single expression cassettes for each enzyme (see also [https://parts.igem.org/Part:BBa_K801073 BBa_K801073], [https://parts.igem.org/Part:BBa_K801074 BBa_K801074] and [https://parts.igem.org/Part:BBa_K801075 BBa_K801075]). It can be directly transformed into competent yeast cells or cloned in an adequate yeast genome integration vector. |
| + | The expression of the enzymes CaXMT1 (''Coffea arabica'' xanthosine-N-methyl-transferase 1, BBa_K801070) and CaDXMT1 (''Coffea arabica'' dimethyl-xanthine N-methyl transferase 1, = caffeine synthase, BBa_K801072) is controlled by the TEF2-promoter (BBa_K801010) and the ADH1-terminator (BBa_K801012) | ||
| + | The expression of CaMXMT1 (''Coffea arabica'' methyl-xanthine N-methyl transferase 1, BBa_K801071) is controlled by the TEF1 promoter (BBa_K319003) and the ADH1-terminator (BBa_K801012) | ||
| + | =Background and principles= | ||
| + | [[Image:TUM12_StructureCaffein.png|thumb|300px|Structure of Caffeine.]] | ||
| − | + | Caffeine is a purine-alkaloid and its biosynthesis occurs in coffee plants and tea plants. Its chemical structure is similar to that of the ribonucleoside adenosine. Hence it can block specific receptors in the hypothalamus. Adenosine binding leads to decreased neurotransmitter-release and therefore decreased neuron activity. This induces sleep and thus avoids overexertion of the brain. Since caffeine antagonizes adenosine and increases neuronal activity, it is used as a means to stay awake. On average, one cup (150 ml) of coffee contains about 50 - 130 mg caffeine and one cup of tea 25 - 90 mg. At higher doses (1g), however, caffeine leads to higher pulse rates and hyperactivity. Moreover, caffeine was shown to decrease the growth of E. Coli and yeast reversibly as of a concentration of 0.1 % by acting as a mutagen (Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972), but previous caffeine synthesis experiments (see below) have only led to a concentration of about 5 µg/g (per g fresh weight of tobacco leaves), so it is not expected to reach critical concentrations and the amounts of caffeine in coffee or tea (leading to physiological effects) is usually a little bit lower. | |
| + | <br/><br/><br/><br/><br/><br/> | ||
| + | ===Usage and Biology=== | ||
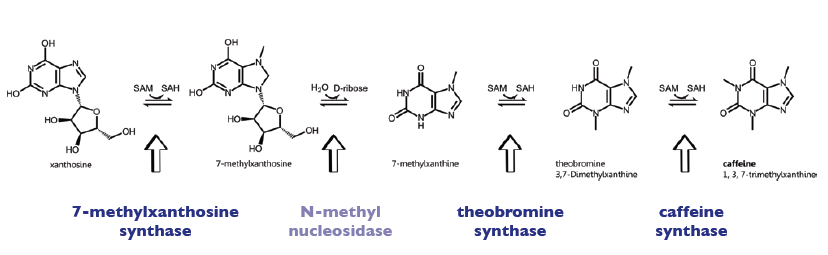
| − | The | + | The enzyme CaXMT1 (xanthosine N-methyltransferase 1 of ''coffea arabica'') catalyses the first reaction step of the caffeine biosynthesis pathway and convertes the sustrate Xanthosine into 7-Methylxanthosine. |
| + | It uses SAM als methyl-donor and is located in the cytoplasm of the plants. | ||
| + | The enzyme CaMXMT1 (7-methylxanthine N-methyltransferase of ''coffea arabica'') catalyses the third reaction step of the caffeine biosynthesis pathway and convertes the sustrate 7-Methylxanthine into 3,7-Dimethylxanthine. | ||
| + | It uses SAM (S-Adenosyl-Methionine) als methyl-donor and is located in the cytoplasm of the plants. | ||
| + | The enzyme CaDXMT1 (3,7-dimethylxanthine N-methyltransferase of ''coffea arabica'') catalyses the fourth reaction step of the caffeine biosynthesis, leading to caffeine. | ||
| + | It uses SAM (S-Adenosyl-Methionine) als methyl-donor and is located in the cytoplasm of the plants. | ||
| + | Furthermore all three exists as homodimer, being also able to form heterodimers with the other enzymes of the caffeine pathway (see [http://www.brenda-enzymes.info Brenda]). | ||
| + | |||
| + | ===Biosynthesis Pathway=== | ||
| + | [[Image:TUM12_Caffeinpathway.png|thumb|left|1000px|Caffeine biosynthesis pathway]] | ||
| + | <br/><br/><br/><br/><br/><br/> | ||
| + | |||
| + | =Characterization= | ||
| + | |||
| + | ===Cloning into pSB1C3=== | ||
| + | [[Image:Erste2.png|thumb|left|100px|Anal. digest]] | ||
| + | |||
| + | In order to submit the enzyme xanthosine methyltransferase (CaXMT1) to the parts.igem, we cloned the generated sequence into pSB1C3, making the system RFC10 compatible. However, since the sequence does not contain any restriction sites of the RFC25 standard (NgoMIV and Age1), RFC25 compatibility can easily be reached without required quick changes by the use of PCR upon usage of appropriate primers. | ||
| + | |||
| + | To check the successful cloning, we performed an analytical digest with XbaI and PstI. | ||
| + | |||
| + | The expected lengths of the fragments were: | ||
| + | <br/><br/> | ||
| + | * Insert (CaXMT1): ca. 6400bp | ||
| + | * Backbone (pSB1C3): ca. 2050 bp | ||
| + | <br/> | ||
| + | The picture on the left shows the analytical digest of BioBrick BBa_K801070 with Xba1 and Pst1, separated by gel-electrophoresis on 1% agarose gel upon usage of ethidium bromide. | ||
| + | ===LC/MS=== | ||
| + | We have been successful in the expression of the three single enzymes making up the caffeine biosynthesis metabolic pathway. Therefore, we used this composite part BBa_K801077 for expression of the enzymes, followed by enzymatic analyses, in order to prove the functionality of our enzymes. After having cloned it into our vector pTUM100, we performed an enzyme assay as described in [Uefuji et al., 2003], with the enzymes having been harvested after 20 hours. The reaction conditions were 27 °C at a pH of 8.0. | ||
| + | Detection of single metabolites of the caffeine biosynthesis pathway was performed by LC/MS analysis, using multiple reaction monitoring (MRM). The picture below shows that we were in fact able to perform an in vitro synthesis of theobromine, which is the immediate precursor of caffeine. However, we failed in the detection of caffeine, because the negative control also showed the corresponding signal with an identical retention time and the amount was not significant, i.e. the e-value was not comparable to the one of the reference. We assumed, that the reason for the failed detection could be the fact, that there was to less produced theobromine in the reaction mixture. Thus, we started another experiment, in which we added theobromine directly to the isolated crude extract. | ||
| + | [[Image:TUM_12Caffein_analysis.jpg|thumb|800px|Used substrate: Xanthosine; A: Reference chromatogram of theobromine; B and C: Chromatograms of two solutions obtained from the enzyme assay; Reacton conditions: T= 27°C, pH= 8.0; Reaction time: 16h; Each 100µl reaction batch consisted of: 50mM Tris/HCl (pH 8.0), 500 µM Xanthosine, 1,5 mM SAM, 200 µM MgCl2, 500 µg protein (crude extract); Reaction products were extracted with 1 ml chloroform, dried at 60°C and subjected to LC/MS analysis after having been resolved in 100µl 70% methanol; ]] | ||
| + | <br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/><br/> | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
| Line 23: | Line 58: | ||
<partinfo>BBa_K801077 parameters</partinfo> | <partinfo>BBa_K801077 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | =References= | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/18068204 Ashihara et al., 2008]] Ashihara, H., Sano, H., and Crozier, A. (2008). Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. ''Phytochemistry'', 69(4):841–56. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/22849837 Franco et al., 2012]] Franco, L., Sánchez, C., Bravo, R., Rodriguez, A., Barriga, C., and Juánez, J. C. (2012). The sedative effects of hops (''humulus lupulus''), a component of beer, on the activity/rest rhythm. ''Acta Physiol Hung'', 99(2):133–9. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/18036626 Kim and Sano, 2008]] Kim, Y.-S. and Sano, H. (2008). Pathogen resistance of transgenic tobacco plants producing caffeine. ''Phytochemistry'', 69(4):882–8. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/16925551 Kuranda et al., 2006]] Kuranda, K., Leberre, V., Sokol, S., Palamarczyk, G., and François, J. (2006). Investigating the caffeine effects in the yeast ''Saccharomyces cerevisiae'' brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. ''Mol Microbiol'', 61(5):1147–66. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1914188/ McCarthy and McCarthy, 2007]] McCarthy, A.A., McCarthy, J.G. (2007). The Structure of Two N-Methyltransferases from the Caffeine Biosynthetic Pathway. ''Plant Physiology'', 144(2):879-889. | ||
| + | *[[http://ci.nii.ac.jp/naid/110006323439/ Negishi et al. (1988)]] Negishi O, Ozawa T and Imagawa H (1988). N-Methyl nucleosidase from tea leaves. ''Agric. Biol. Chem.'' 52: 169–175. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/12746542 Uefuji et al., 2003]] Uefuji, H., Ogita, S., Yamaguchi, Y., Koizumi, N., and Sano, H. (2003). Molecular cloning and functional characterization of three distinct n-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. ''Plant Physiol'', 132(1):372–80. | ||
| + | *[[http://www.ncbi.nlm.nih.gov/pubmed/16247553 Uefuji et al., 2005]] Uefuji, H., Tatsumi, Y., Morimoto, M., Kaothien-Nakayama, P., Ogita, S., and Sano, H. (2005). Caffeine production in tobacco plants by simultaneous expression of three coffee n-methyltrasferases and its potential as a pest repellant. ''Plant Mol Biol'', 59(2):221–7. | ||
Latest revision as of 19:49, 26 October 2012
Caffeine Synthesis Pathway pTEF2-XMT1-tADH1-pTEF1-MXMT1-tADH1-pTEF2-DXMT1-tADH1
This BioBrick is the final "caffeine synthesis device". It contains all three necessary enzymes: CaXMT1, CaMXMT1 and CaDXMT1, i.e. it is made up of the three single expression cassettes for each enzyme (see also BBa_K801073, BBa_K801074 and BBa_K801075). It can be directly transformed into competent yeast cells or cloned in an adequate yeast genome integration vector. The expression of the enzymes CaXMT1 (Coffea arabica xanthosine-N-methyl-transferase 1, BBa_K801070) and CaDXMT1 (Coffea arabica dimethyl-xanthine N-methyl transferase 1, = caffeine synthase, BBa_K801072) is controlled by the TEF2-promoter (BBa_K801010) and the ADH1-terminator (BBa_K801012) The expression of CaMXMT1 (Coffea arabica methyl-xanthine N-methyl transferase 1, BBa_K801071) is controlled by the TEF1 promoter (BBa_K319003) and the ADH1-terminator (BBa_K801012)
Background and principles
Caffeine is a purine-alkaloid and its biosynthesis occurs in coffee plants and tea plants. Its chemical structure is similar to that of the ribonucleoside adenosine. Hence it can block specific receptors in the hypothalamus. Adenosine binding leads to decreased neurotransmitter-release and therefore decreased neuron activity. This induces sleep and thus avoids overexertion of the brain. Since caffeine antagonizes adenosine and increases neuronal activity, it is used as a means to stay awake. On average, one cup (150 ml) of coffee contains about 50 - 130 mg caffeine and one cup of tea 25 - 90 mg. At higher doses (1g), however, caffeine leads to higher pulse rates and hyperactivity. Moreover, caffeine was shown to decrease the growth of E. Coli and yeast reversibly as of a concentration of 0.1 % by acting as a mutagen (Putrament et al., On the Specificity of Caffeine Effects, MGG, 1972), but previous caffeine synthesis experiments (see below) have only led to a concentration of about 5 µg/g (per g fresh weight of tobacco leaves), so it is not expected to reach critical concentrations and the amounts of caffeine in coffee or tea (leading to physiological effects) is usually a little bit lower.
Usage and Biology
The enzyme CaXMT1 (xanthosine N-methyltransferase 1 of coffea arabica) catalyses the first reaction step of the caffeine biosynthesis pathway and convertes the sustrate Xanthosine into 7-Methylxanthosine. It uses SAM als methyl-donor and is located in the cytoplasm of the plants. The enzyme CaMXMT1 (7-methylxanthine N-methyltransferase of coffea arabica) catalyses the third reaction step of the caffeine biosynthesis pathway and convertes the sustrate 7-Methylxanthine into 3,7-Dimethylxanthine. It uses SAM (S-Adenosyl-Methionine) als methyl-donor and is located in the cytoplasm of the plants. The enzyme CaDXMT1 (3,7-dimethylxanthine N-methyltransferase of coffea arabica) catalyses the fourth reaction step of the caffeine biosynthesis, leading to caffeine. It uses SAM (S-Adenosyl-Methionine) als methyl-donor and is located in the cytoplasm of the plants. Furthermore all three exists as homodimer, being also able to form heterodimers with the other enzymes of the caffeine pathway (see [http://www.brenda-enzymes.info Brenda]).
Biosynthesis Pathway
Characterization
Cloning into pSB1C3
In order to submit the enzyme xanthosine methyltransferase (CaXMT1) to the parts.igem, we cloned the generated sequence into pSB1C3, making the system RFC10 compatible. However, since the sequence does not contain any restriction sites of the RFC25 standard (NgoMIV and Age1), RFC25 compatibility can easily be reached without required quick changes by the use of PCR upon usage of appropriate primers.
To check the successful cloning, we performed an analytical digest with XbaI and PstI.
The expected lengths of the fragments were:
- Insert (CaXMT1): ca. 6400bp
- Backbone (pSB1C3): ca. 2050 bp
The picture on the left shows the analytical digest of BioBrick BBa_K801070 with Xba1 and Pst1, separated by gel-electrophoresis on 1% agarose gel upon usage of ethidium bromide.
LC/MS
We have been successful in the expression of the three single enzymes making up the caffeine biosynthesis metabolic pathway. Therefore, we used this composite part BBa_K801077 for expression of the enzymes, followed by enzymatic analyses, in order to prove the functionality of our enzymes. After having cloned it into our vector pTUM100, we performed an enzyme assay as described in [Uefuji et al., 2003], with the enzymes having been harvested after 20 hours. The reaction conditions were 27 °C at a pH of 8.0. Detection of single metabolites of the caffeine biosynthesis pathway was performed by LC/MS analysis, using multiple reaction monitoring (MRM). The picture below shows that we were in fact able to perform an in vitro synthesis of theobromine, which is the immediate precursor of caffeine. However, we failed in the detection of caffeine, because the negative control also showed the corresponding signal with an identical retention time and the amount was not significant, i.e. the e-value was not comparable to the one of the reference. We assumed, that the reason for the failed detection could be the fact, that there was to less produced theobromine in the reaction mixture. Thus, we started another experiment, in which we added theobromine directly to the isolated crude extract.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 3613
Illegal BglII site found at 5829
Illegal BglII site found at 5925 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 138
Illegal NgoMIV site found at 4317 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2385
References
- http://www.ncbi.nlm.nih.gov/pubmed/18068204 Ashihara et al., 2008 Ashihara, H., Sano, H., and Crozier, A. (2008). Caffeine and related purine alkaloids: biosynthesis, catabolism, function and genetic engineering. Phytochemistry, 69(4):841–56.
- http://www.ncbi.nlm.nih.gov/pubmed/22849837 Franco et al., 2012 Franco, L., Sánchez, C., Bravo, R., Rodriguez, A., Barriga, C., and Juánez, J. C. (2012). The sedative effects of hops (humulus lupulus), a component of beer, on the activity/rest rhythm. Acta Physiol Hung, 99(2):133–9.
- http://www.ncbi.nlm.nih.gov/pubmed/18036626 Kim and Sano, 2008 Kim, Y.-S. and Sano, H. (2008). Pathogen resistance of transgenic tobacco plants producing caffeine. Phytochemistry, 69(4):882–8.
- http://www.ncbi.nlm.nih.gov/pubmed/16925551 Kuranda et al., 2006 Kuranda, K., Leberre, V., Sokol, S., Palamarczyk, G., and François, J. (2006). Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol Microbiol, 61(5):1147–66.
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1914188/ McCarthy and McCarthy, 2007 McCarthy, A.A., McCarthy, J.G. (2007). The Structure of Two N-Methyltransferases from the Caffeine Biosynthetic Pathway. Plant Physiology, 144(2):879-889.
- http://ci.nii.ac.jp/naid/110006323439/ Negishi et al. (1988) Negishi O, Ozawa T and Imagawa H (1988). N-Methyl nucleosidase from tea leaves. Agric. Biol. Chem. 52: 169–175.
- http://www.ncbi.nlm.nih.gov/pubmed/12746542 Uefuji et al., 2003 Uefuji, H., Ogita, S., Yamaguchi, Y., Koizumi, N., and Sano, H. (2003). Molecular cloning and functional characterization of three distinct n-methyltransferases involved in the caffeine biosynthetic pathway in coffee plants. Plant Physiol, 132(1):372–80.
- http://www.ncbi.nlm.nih.gov/pubmed/16247553 Uefuji et al., 2005 Uefuji, H., Tatsumi, Y., Morimoto, M., Kaothien-Nakayama, P., Ogita, S., and Sano, H. (2005). Caffeine production in tobacco plants by simultaneous expression of three coffee n-methyltrasferases and its potential as a pest repellant. Plant Mol Biol, 59(2):221–7.