Difference between revisions of "Part:BBa K823038"
(→Characterization: TU_Dresden 2019) |
|||
| (24 intermediate revisions by 4 users not shown) | |||
| Line 28: | Line 28: | ||
| − | Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''BacillusB'''''io'''B'''rick'''B'''ox]. | + | Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''BacillusB'''''io'''B'''rick'''B'''ox]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The ''Bacillus'' BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with ''Bacillus subtilis''] by Radeck ''et al.''. |
| + | |||
| + | ===Evaluation=== | ||
| + | <p align="justify"> | ||
| + | All 5 epitope tags were fused C- and N-terminally to GFP using the NgoMIV and AgeI restriction sites. These constructs were expressed in ''Bacillus subtils'' using [https://parts.igem.org/Part:BBa_K823026 pSB<sub>Bs</sub>0K-P<sub>spac</sub>]. This vector did not need to be induced by IPTG due to a premature stop codon in the lacI gene. | ||
| + | |||
| + | |||
| + | </p> | ||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[Image:LMU-Western_Blot_Tags.png|400px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
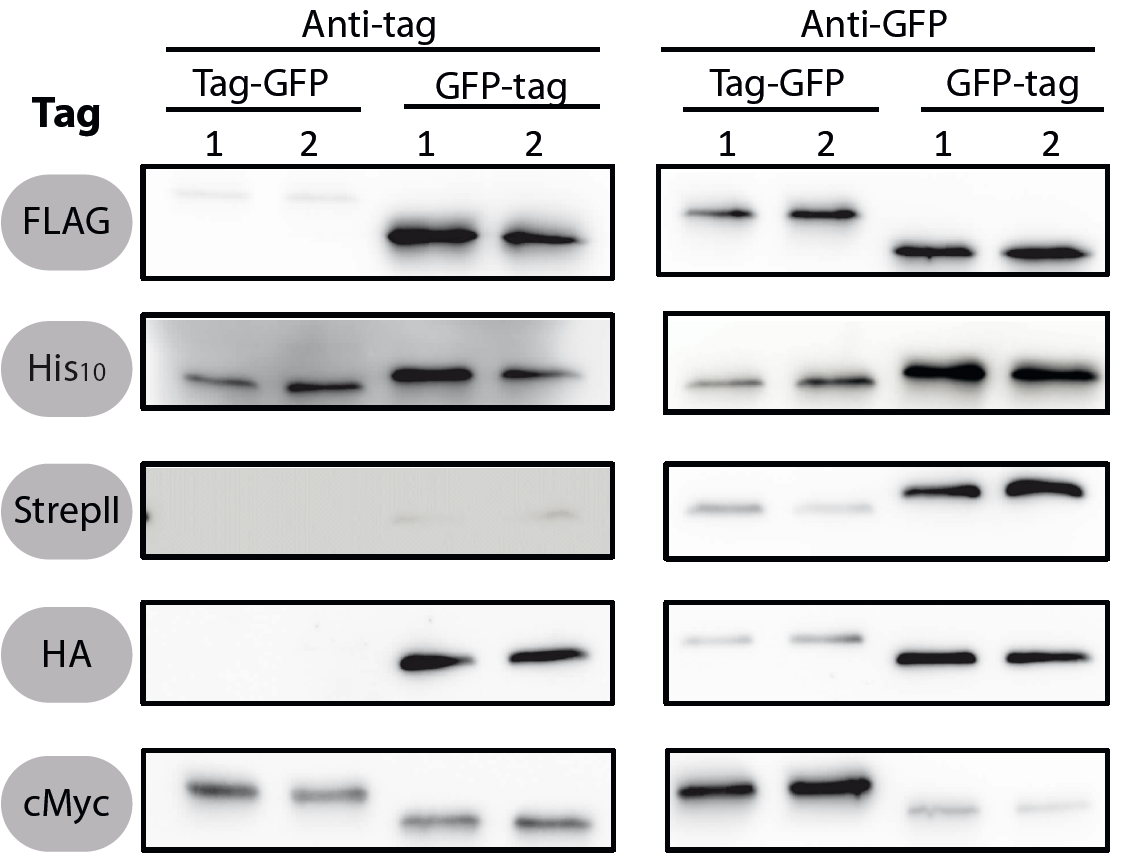
| + | <font color="#000000"; size="2"><p align="justify"> Fig. 1: Western blots of N- and C-terminal fusions of each tag to GFP, using the strains TMB1920 (Flag-gfp), TMB1921 (gfp-Flag), TMB1922 (HA-gfp), TMB1923 (gfp-HA), TMB1924 (cMyc-gfp), TMB1925 (gfp-cMyc), TMB1926 (His-gfp), TMB1927 (gfp-His), TMB1928 (StrepII-gfp) and TMB1929 (gfp-StrepII). For each construct, two independent clones were tested with epitope tag- and GFP-specific antibodies as a positive control. | ||
| + | |||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | ===Methods=== | ||
| + | |||
| + | |||
| + | To verify the functionality of the epitope tags, Western blot analyses of the strains TMB1920-TMB1929 were performed. LB medium (15 ml) was inoculated 1:100 from overnight culture and grown at 37°C and 200 rpm to OD600 ~ 0.5. Of this, 10 ml were harvested by centrifugation (8000 × g, 5 min) and the pellets stored at -20°C. Pellets were resuspended in 1 ml disruption buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl) and lysed by sonication. Samples (12 μl of lysate) were loaded per lane on two 12.5% SDS-polyacrylamide gels and SDS-PAGE was performed according standard procedure [60]. One gel was stained with colloidal coomassie, the other one was used for protein transfer to a PVDF membrane (Merck Millipore, Billerica, MA, USA) by submerged blotting procedure (Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA)). After protein transfer, the membranes were treated with the following antibodies and conditions. Detailed protocols can be found [http://www.jbioleng.org/content/7/1/29/suppl/S3 here]. | ||
| + | |||
| + | |||
| + | ''GFP'' | ||
| + | |||
| + | Probing with primary antibodies takes place with rabbit anti-GFP antibodies (1:3000, Epitomics, No. 1533). Horseradish-peroxidase (HRP)-conjugated anti-rabbit antibodies (1:2000, Promega, W401B) were used as secondary antibody. Hybridization of both antibodies was carried out in Blotto-buffer (2.5% (w/v) skim milk powder, 1 × TBS (50 mM Tris–HCl pH 7.6, 0.15 M NaCl)). | ||
| + | |||
| + | |||
| + | ''StrepII'' | ||
| + | |||
| + | Strep-Tactin-HRP conjugate (IBA, Strep-Tactin-HRP conjugate, No. 2-1502-001) 1:100 in 1 × PBS (4 mM KH2PO4; 16 mM Na2HPO4; 115 mM NaCl) with 0.1% (w/v) Tween20 was used. | ||
| + | |||
| + | |||
| + | Chemiluminescence signals were detected after addition of the HRP-substrate Ace Glow (Peqlab, Erlangen, Germany) using a Fusion<sup>TM</sup> imaging system (Peqlab). | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | === Characterization: [https://2019.igem.org/Team:TU_Dresden TU_Dresden 2019] === | ||
| + | |||
| + | ==== Strep-tag column purification ==== | ||
| + | |||
| + | The Team TU Dresden 2019 used this Biobrick for purification purposes using the “Expression and purification of proteins using Strep-Tactin” protocol of IBA Lifescience [1]. We tried to purify many of our different BioBricks via this method, in order to characterize the proper function of the Strep-tag for purification purposes. However, we were not able to obtain purified constructs, meaning that this Strep-tag should rather be used for Western Blotting instead of for purification. Nevertheless, the different BioBricks that we tried to purify with this Strep-tag have shown to be working properly (see their Registries: [https://parts.igem.org/Part:BBa_K3037003 BBa_K3037003,] [https://parts.igem.org/Part:BBa_K3037003 BBa_K3037009)] | ||
| + | |||
| + | The results of the different purification tests are shown in the following gels: | ||
| + | |||
| + | ===== a) Purification of our Full Construct ([https://parts.igem.org/Part:BBa_K3037003 BBa_K3037003)] ===== | ||
| + | |||
| + | [[File:T--TU_Dresden--Strep-tag_purification_BBa_3037003.png|center|400px|thumb|none|Fig.2 SDS-PAGE showing the unsuccessful purification of our Full Construct [https://parts.igem.org/Part:BBa_K3037003 BBa_K3037003]. The elution fractions do not contain our construct of interest. ]] | ||
| + | |||
| + | |||
| + | The purification of this construct in Fig. 2 was repeated again by using the same protocol, but this time a successful MBP-tag purification was performed first (see the registry page of our Full Construct for more details [https://parts.igem.org/Part:BBa_K3037003 BBa_K3037003]). As shown in the gel of Fig. 3, the Strep-tag column purification did not work. | ||
| + | [[File:Strep_purification_2.png|center|400px|thumb|none|Fig. 3 SDS-PAGE showing the unsuccessful purification of our Full Construct. The elution fractions do not contain our construct of interest, although this construct was already successfully purified via its MBP. A big part of our Full Construct seemed to be went through the column, since it is visible in the cell lysate, flowthrough fraction washing step between 180 and 245 kDa.]] | ||
| + | |||
| + | |||
| + | ===== b) Purification of HRP-Strep ([https://parts.igem.org/Part:BBa_K3037009 BBa_K3037009)] ===== | ||
| + | |||
| + | The protocol used was “Expression and purification of proteins using Strep-Tactin” of IBA Lifescience [1] preparing the same buffers described in this protocols without EDTA to not harm the activity of HRP. From the results of the purifications we concluded that the Strep-tag is not working properly for column purification and should therefore only be used for Western Blots. | ||
| + | |||
| + | [[File:T--TU_Dresden--Strep-tag_purification_BBa_3037009.png|center|400px|thumb|none|Fig. 4 SDS-PAGE showing the unsuccessful purification of the HRP-Strep construct. The elution fractions do not contain our construct of interest.]] | ||
| + | |||
| + | |||
| + | ===== c) Extracting the Strep column and loading it into a SDS-page ===== | ||
| + | |||
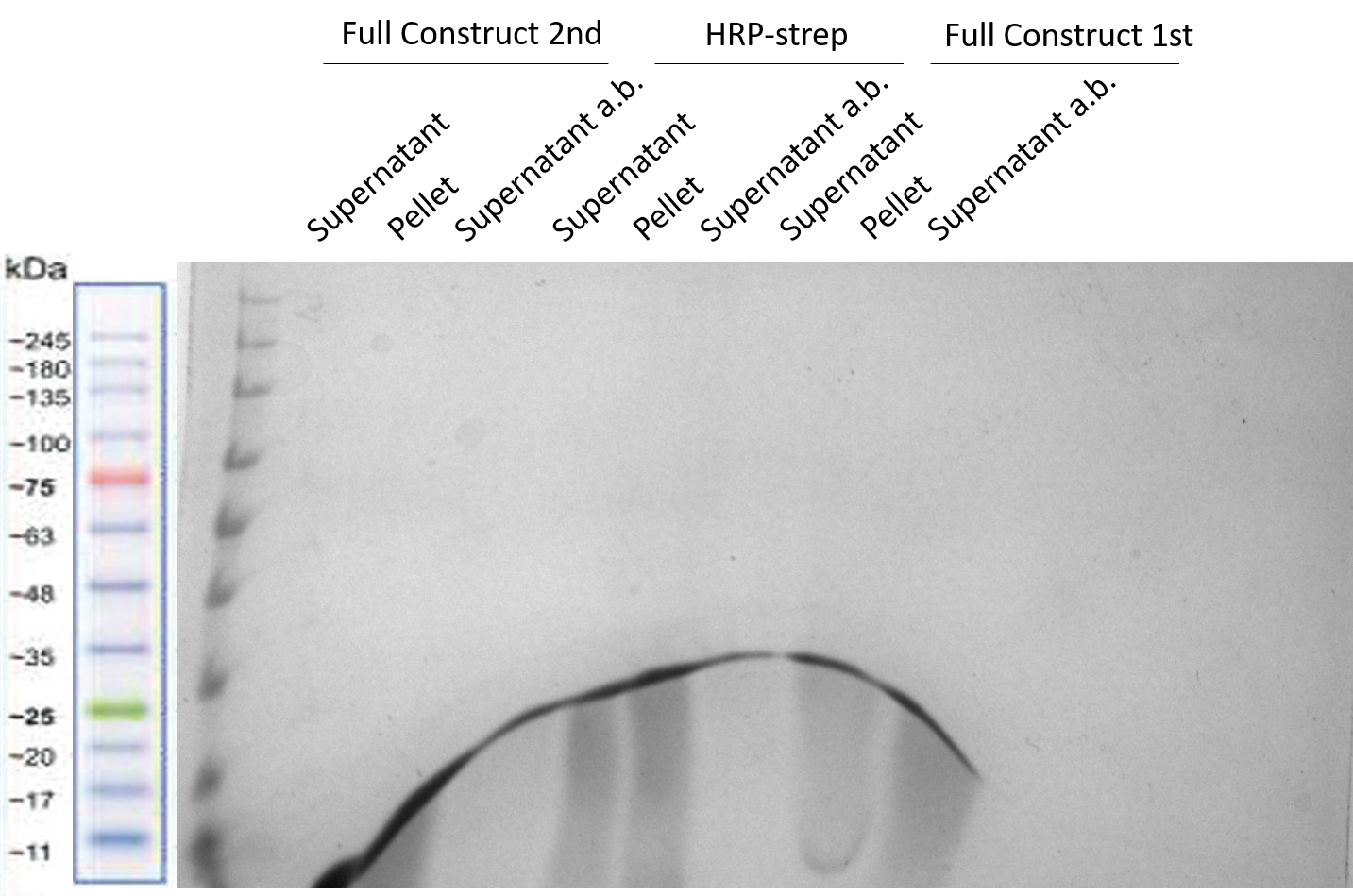
| + | In order to be able to confirm that the different constructs mentioned above did not bind to the Strep, we decided to extract the Strep from the column and load it into a SDS-page. We were doubting if the proteins could have bound to the Strep inside of the column but were not eluted properly. That is why we extracted the Strep from the three different columns, resuspended it in W-buffer from [1] and added SDS-loading gel to it. Three different samples of each column were prepared (supernatant and pellet of centrifuging the Strep samples) and finally a third sample of the supernatant after adding SDS. In Figure 5, these three samples each of HRP-strep and Full Construct (1st and 2nd trial)are shown. | ||
| + | |||
| + | |||
| + | [[File:T--TU_Dresden--emptystrep.png|center|400px|thumb|none|Fig. 5 SDS-PAGE containing the Strep-tags extracted from the three different purification columns: Full Construct (2nd trial), HRP-strep and Full Construct (1st trial).]] | ||
| + | |||
| + | It can be clearly seen, that the proteins were not bound to the Strep-tags of the columns either. This suggests that indeed, the Strep is not working properly for purification columns. As it was already clearly seen in Figure 2, most part of our proteins of interest just flowed through. | ||
| + | |||
| + | ===== Conclusions ===== | ||
| + | |||
| + | From the results of these purifications in Fig. 2 to 5, we can conclude that the Strep-tag does not properly work for column purification and should rather be used for Western Blotting. | ||
| + | |||
| + | ===== References ===== | ||
| + | [1] https://www.iba-lifesciences.com/isotope/2/2-3206-100-Manual-Twin--Strep-tag.pdf | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 01:59, 22 October 2019
Strep-tag (Freiburg standard+RBS)
Streptavidin - tag with RBS in Freiburg standard.
Find out more about the design of our prefix with ribosome binding site.
prefix:GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix:ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
The Strep-tag is a mimicry peptide of biotin which binds to Streptavidin ([http://www.sciencedirect.com/science/article/pii/S1050386299000339 Skerra, A. and Schmidt, T.G.M. (1999)]). Its sequence is WSHPQFEK. It can be used for protein purification, immobilisation with Streptavidin or Strep-tactin ([http://www.ncbi.nlm.nih.gov/pubmed/9415448 Voss, S. and Skerra, A. (1997)]) or detection with Strep-tactin or antibodies.
This is a part created by the LMU-Munich 2012 team. We added five tags to the registry, all in the Freiburg standard for N-and C-terminal fusions:
- Strep - tag
Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks BacillusBioBrickBox]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
Evaluation
All 5 epitope tags were fused C- and N-terminally to GFP using the NgoMIV and AgeI restriction sites. These constructs were expressed in Bacillus subtils using pSBBs0K-Pspac. This vector did not need to be induced by IPTG due to a premature stop codon in the lacI gene.
|
Methods
To verify the functionality of the epitope tags, Western blot analyses of the strains TMB1920-TMB1929 were performed. LB medium (15 ml) was inoculated 1:100 from overnight culture and grown at 37°C and 200 rpm to OD600 ~ 0.5. Of this, 10 ml were harvested by centrifugation (8000 × g, 5 min) and the pellets stored at -20°C. Pellets were resuspended in 1 ml disruption buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl) and lysed by sonication. Samples (12 μl of lysate) were loaded per lane on two 12.5% SDS-polyacrylamide gels and SDS-PAGE was performed according standard procedure [60]. One gel was stained with colloidal coomassie, the other one was used for protein transfer to a PVDF membrane (Merck Millipore, Billerica, MA, USA) by submerged blotting procedure (Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA)). After protein transfer, the membranes were treated with the following antibodies and conditions. Detailed protocols can be found [http://www.jbioleng.org/content/7/1/29/suppl/S3 here].
GFP
Probing with primary antibodies takes place with rabbit anti-GFP antibodies (1:3000, Epitomics, No. 1533). Horseradish-peroxidase (HRP)-conjugated anti-rabbit antibodies (1:2000, Promega, W401B) were used as secondary antibody. Hybridization of both antibodies was carried out in Blotto-buffer (2.5% (w/v) skim milk powder, 1 × TBS (50 mM Tris–HCl pH 7.6, 0.15 M NaCl)).
StrepII
Strep-Tactin-HRP conjugate (IBA, Strep-Tactin-HRP conjugate, No. 2-1502-001) 1:100 in 1 × PBS (4 mM KH2PO4; 16 mM Na2HPO4; 115 mM NaCl) with 0.1% (w/v) Tween20 was used.
Chemiluminescence signals were detected after addition of the HRP-substrate Ace Glow (Peqlab, Erlangen, Germany) using a FusionTM imaging system (Peqlab).
Characterization: TU_Dresden 2019
Strep-tag column purification
The Team TU Dresden 2019 used this Biobrick for purification purposes using the “Expression and purification of proteins using Strep-Tactin” protocol of IBA Lifescience [1]. We tried to purify many of our different BioBricks via this method, in order to characterize the proper function of the Strep-tag for purification purposes. However, we were not able to obtain purified constructs, meaning that this Strep-tag should rather be used for Western Blotting instead of for purification. Nevertheless, the different BioBricks that we tried to purify with this Strep-tag have shown to be working properly (see their Registries: BBa_K3037003, BBa_K3037009)
The results of the different purification tests are shown in the following gels:
a) Purification of our Full Construct (BBa_K3037003)
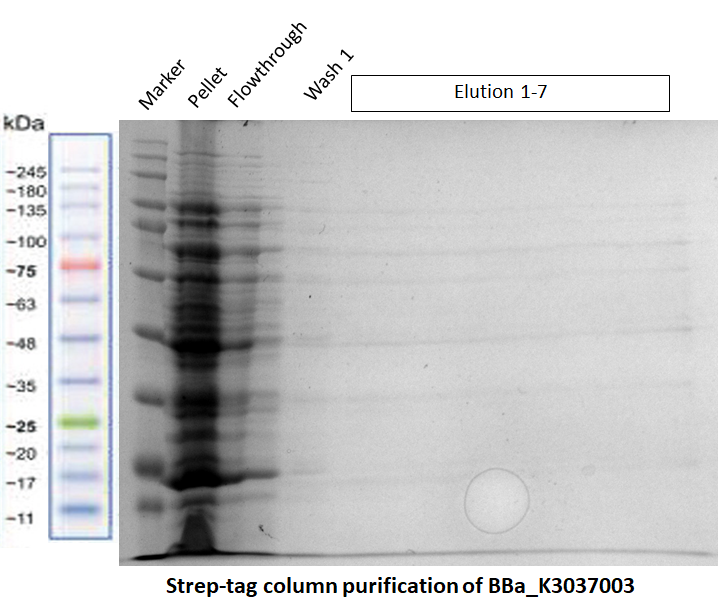
The purification of this construct in Fig. 2 was repeated again by using the same protocol, but this time a successful MBP-tag purification was performed first (see the registry page of our Full Construct for more details BBa_K3037003). As shown in the gel of Fig. 3, the Strep-tag column purification did not work.
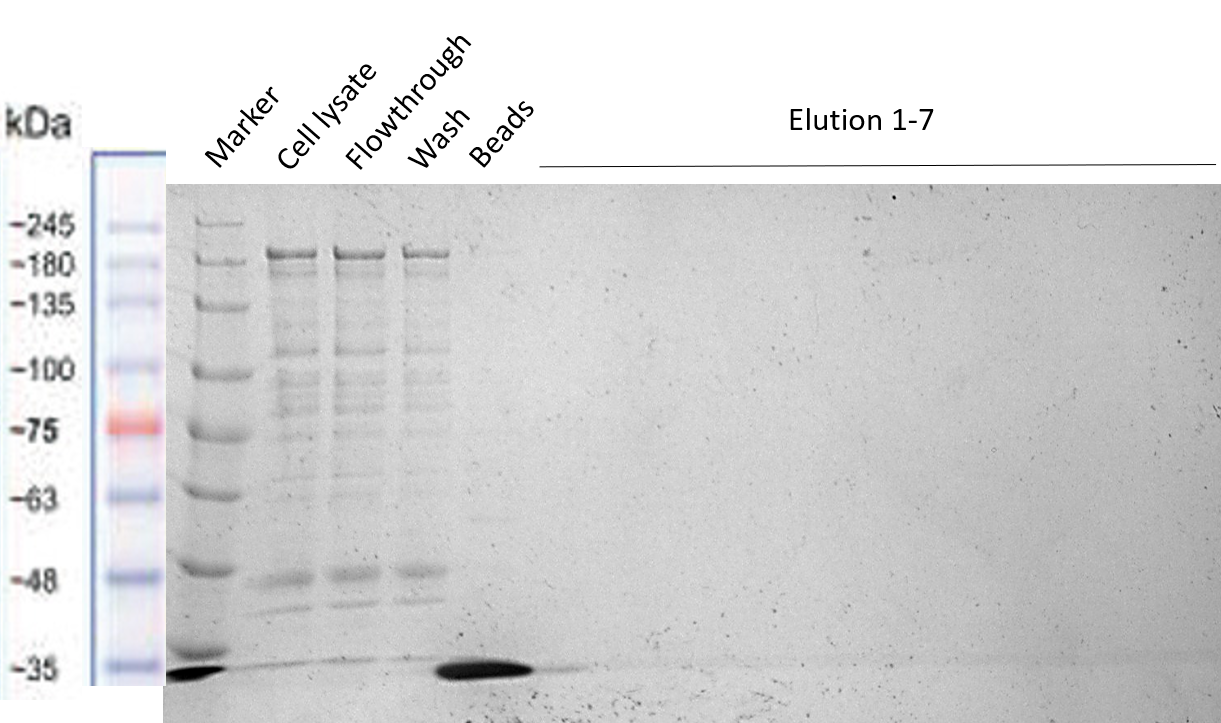
b) Purification of HRP-Strep (BBa_K3037009)
The protocol used was “Expression and purification of proteins using Strep-Tactin” of IBA Lifescience [1] preparing the same buffers described in this protocols without EDTA to not harm the activity of HRP. From the results of the purifications we concluded that the Strep-tag is not working properly for column purification and should therefore only be used for Western Blots.
c) Extracting the Strep column and loading it into a SDS-page
In order to be able to confirm that the different constructs mentioned above did not bind to the Strep, we decided to extract the Strep from the column and load it into a SDS-page. We were doubting if the proteins could have bound to the Strep inside of the column but were not eluted properly. That is why we extracted the Strep from the three different columns, resuspended it in W-buffer from [1] and added SDS-loading gel to it. Three different samples of each column were prepared (supernatant and pellet of centrifuging the Strep samples) and finally a third sample of the supernatant after adding SDS. In Figure 5, these three samples each of HRP-strep and Full Construct (1st and 2nd trial)are shown.
It can be clearly seen, that the proteins were not bound to the Strep-tags of the columns either. This suggests that indeed, the Strep is not working properly for purification columns. As it was already clearly seen in Figure 2, most part of our proteins of interest just flowed through.
Conclusions
From the results of these purifications in Fig. 2 to 5, we can conclude that the Strep-tag does not properly work for column purification and should rather be used for Western Blotting.
References
[1] https://www.iba-lifesciences.com/isotope/2/2-3206-100-Manual-Twin--Strep-tag.pdf
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]