Difference between revisions of "Part:BBa K863005:Experience"
Sednanalien (Talk | contribs) (→User Reviews) |
(→User Reviews) |
||
| (6 intermediate revisions by 4 users not shown) | |||
| Line 18: | Line 18: | ||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
<!-- DON'T DELETE --><partinfo>BBa_K863005 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_K863005 EndReviews</partinfo> | ||
| + | |||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K863005 AddReview 5</partinfo> | ||
| + | <I>Slovenia_HS</I> | ||
| + | |width='60%' valign='top'| | ||
| + | Slovenia HS characterized this part in 2015. | ||
| + | |||
| + | Escherichia coli BL21 (DE3) bacteria were transformed with the expression plasmids (BBa_K863005 and BBa_K863010) and grown in 10 ml at 37 °C in LBC medium overnight. To express both recombinant proteins, 10 ml of overnight cultures shaker cultures were grown at 37 °C in LB broth supplemented with 30 µg/ml chloramphenicol and shaking with 225 rpm. Expression of the recombinant protein was induced by addition of IPTG to a final concentration of 1 mM, when the cell density reached an OD600 of 0.8. After induction, cells were grown for 5 h and then collected by centrifugation at 6000g for 10 min. The cell pellet collected from 400 ml of bacterial culture was resuspended in 20 ml of resuspension buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole) and sonified 3 × 6 min on ice. Following centrifugation at 30 000 × g for 10 min to remove insoluble debris, the supernatant was applied to a Ni-NTA Superflow Cartridge (Qiagen) connected to ÄKTA FPLC system, washed with the resuspension buffer and eluted in the same buffer, but containing 250 mM imidazole. The peak fractions were collected and 15 µl of each fraction was collected and resolved on 12 % SDS-PAGE. | ||
| + | |||
| + | |||
| + | https://static.igem.org/mediawiki/2015/thumb/3/31/Lakaze.jpeg/293px-Lakaze.jpeg | ||
| + | |||
| + | |||
| + | We found that while BBa_K863005 shows excellent activity, BBa_K863010 shows no activity under the same conditions. | ||
| + | |}; | ||
{|width='80%' style='border:1px solid gray' | {|width='80%' style='border:1px solid gray' | ||
| Line 25: | Line 43: | ||
<I>University College London 2012</I> | <I>University College London 2012</I> | ||
|width='60%' valign='top'| | |width='60%' valign='top'| | ||
| − | As part of our collaboration with Bielefeld-Germany, we have characterised their Laccase BioBrick utilising a Syringaldazine assay to determine their Laccase activity. Syringaldazine is oxidised by Laccase, causing a colourmetric change that can be observed by a spectrophotometer at 530nm. As seen in the graph below, the highest rate of Laccase production is in transformed BL21 cells - this is to be expected due to the T7 promoter, which requires T7 RNA polymerase that BL21 produces. However, expression in W3110, which does not produce T7 RNA polymerase, is still elevated above that of untransformed ''E. coli'', though not at the level of expression of transformed BL21. We hence conclude that Bielefeld-Germany's Laccase BioBrick is working as expected. | + | As part of our collaboration with Bielefeld-Germany, we have characterised their Laccase BioBrick utilising a Syringaldazine assay to determine their Laccase activity. Syringaldazine is oxidised by Laccase, causing a colourmetric change that can be observed by a spectrophotometer at 530nm. As seen in the graph below, the highest rate of Laccase production is in transformed BL21 cells - this is to be expected due to the T7 promoter, which requires the T7 RNA polymerase that BL21 produces. However, expression in transformed W3110, which does not produce T7 RNA polymerase, is still elevated above that of untransformed ''E. coli'', though not at the level of expression of transformed BL21. We hence conclude that Bielefeld-Germany's Laccase BioBrick is working as expected. |
[[Image:UniversityCollegeLondon_CollaborationLaccase.png|800px|centre]] | [[Image:UniversityCollegeLondon_CollaborationLaccase.png|800px|centre]] | ||
| + | |}; | ||
| + | |||
| + | |||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_K863005 AddReview 5</partinfo> | ||
| + | <I>CHINA_CD_UESTC</I> | ||
| + | |width='60%' valign='top'| | ||
| + | We transferred the two vectors (BBa_K1779200 and BBa_K1779204) into ''Escherichia coli BL21 (DE3)'' respectively and grown in 20ml at 37°C for 3 days. Then we broke the cell with Ultrasonic Cell Disruptor and got the mixture. So the mixture contained the phusion proteins we could use to our enzymatic biofuel cell (EBFC). To detect our phusion proteins' catalytic activity, we used the ABTS method to deal with it. | ||
| + | https://static.igem.org/mediawiki/2015/e/e5/CHINA_CD_UESTC_RESULTfin05.png | ||
| + | '''Figure 1. The color of supernate and activity of laccase. (A)''' The higher concentration of laccase showed redder. '''(B)''' The activity of RFP+Laccase. Ultrasonic Cell Disruptor to crush the bacterium in ice-bath. Collect the Supernatant and detect the activity of Laccase by ABTS method. The 1mL supernate equal to the 5mL bacterium liquid which were cultivated for different times. | ||
| + | https://static.igem.org/mediawiki/2015/8/8b/CHINA_CD_UESTC_RESULTfin06.png | ||
| + | '''Figure 2. The color of supernate and activity of laccase. (A)''' The left is ''Escherichia coli BL21(DE3)'' untransformed and the right is ''Escherichia coli BL21(DE3)'' transformed with piGEM-WRL. '''(B)''' The activity of MamW + RFP + Laccase. | ||
|}; | |}; | ||
Latest revision as of 02:59, 18 September 2015
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K863005
User Reviews
UNIQ47525b569b6edd62-partinfo-00000000-QINU UNIQ47525b569b6edd62-partinfo-00000001-QINU
|
•••••
Slovenia_HS |
Slovenia HS characterized this part in 2015. Escherichia coli BL21 (DE3) bacteria were transformed with the expression plasmids (BBa_K863005 and BBa_K863010) and grown in 10 ml at 37 °C in LBC medium overnight. To express both recombinant proteins, 10 ml of overnight cultures shaker cultures were grown at 37 °C in LB broth supplemented with 30 µg/ml chloramphenicol and shaking with 225 rpm. Expression of the recombinant protein was induced by addition of IPTG to a final concentration of 1 mM, when the cell density reached an OD600 of 0.8. After induction, cells were grown for 5 h and then collected by centrifugation at 6000g for 10 min. The cell pellet collected from 400 ml of bacterial culture was resuspended in 20 ml of resuspension buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 20 mM imidazole) and sonified 3 × 6 min on ice. Following centrifugation at 30 000 × g for 10 min to remove insoluble debris, the supernatant was applied to a Ni-NTA Superflow Cartridge (Qiagen) connected to ÄKTA FPLC system, washed with the resuspension buffer and eluted in the same buffer, but containing 250 mM imidazole. The peak fractions were collected and 15 µl of each fraction was collected and resolved on 12 % SDS-PAGE.
|
|
•••••
University College London 2012 |
As part of our collaboration with Bielefeld-Germany, we have characterised their Laccase BioBrick utilising a Syringaldazine assay to determine their Laccase activity. Syringaldazine is oxidised by Laccase, causing a colourmetric change that can be observed by a spectrophotometer at 530nm. As seen in the graph below, the highest rate of Laccase production is in transformed BL21 cells - this is to be expected due to the T7 promoter, which requires the T7 RNA polymerase that BL21 produces. However, expression in transformed W3110, which does not produce T7 RNA polymerase, is still elevated above that of untransformed E. coli, though not at the level of expression of transformed BL21. We hence conclude that Bielefeld-Germany's Laccase BioBrick is working as expected. |
|
•••••
CHINA_CD_UESTC |
We transferred the two vectors (BBa_K1779200 and BBa_K1779204) into Escherichia coli BL21 (DE3) respectively and grown in 20ml at 37°C for 3 days. Then we broke the cell with Ultrasonic Cell Disruptor and got the mixture. So the mixture contained the phusion proteins we could use to our enzymatic biofuel cell (EBFC). To detect our phusion proteins' catalytic activity, we used the ABTS method to deal with it.
|


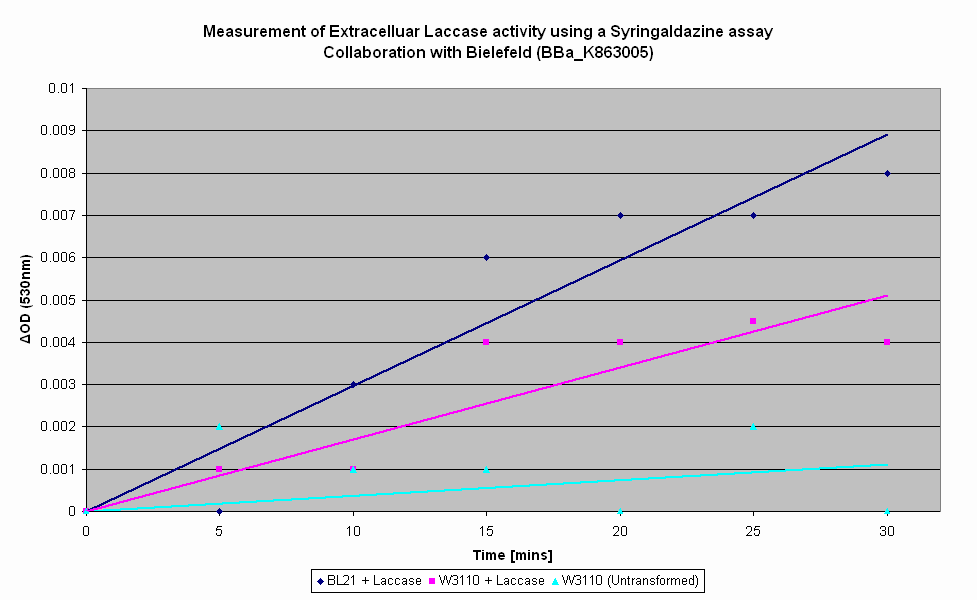
 Figure 1. The color of supernate and activity of laccase. (A) The higher concentration of laccase showed redder. (B) The activity of RFP+Laccase. Ultrasonic Cell Disruptor to crush the bacterium in ice-bath. Collect the Supernatant and detect the activity of Laccase by ABTS method. The 1mL supernate equal to the 5mL bacterium liquid which were cultivated for different times.
Figure 1. The color of supernate and activity of laccase. (A) The higher concentration of laccase showed redder. (B) The activity of RFP+Laccase. Ultrasonic Cell Disruptor to crush the bacterium in ice-bath. Collect the Supernatant and detect the activity of Laccase by ABTS method. The 1mL supernate equal to the 5mL bacterium liquid which were cultivated for different times.
 Figure 2. The color of supernate and activity of laccase. (A) The left is Escherichia coli BL21(DE3) untransformed and the right is Escherichia coli BL21(DE3) transformed with piGEM-WRL. (B) The activity of MamW + RFP + Laccase.
Figure 2. The color of supernate and activity of laccase. (A) The left is Escherichia coli BL21(DE3) untransformed and the right is Escherichia coli BL21(DE3) transformed with piGEM-WRL. (B) The activity of MamW + RFP + Laccase.