Difference between revisions of "Part:BBa J176005"
| (2 intermediate revisions by one other user not shown) | |||
| Line 8: | Line 8: | ||
[[Image:Haynes_discosomasp.png|200px|thumb|The mushroom coral Discosoma sp. is the source of monomeric RFP.]]<br> | [[Image:Haynes_discosomasp.png|200px|thumb|The mushroom coral Discosoma sp. is the source of monomeric RFP.]]<br> | ||
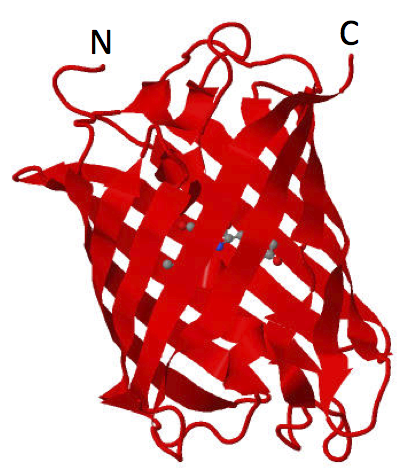
| − | [[Image:Haynes_mCherry_PDB2H5Q.png|200px|thumb|mCherry is a mutant derivative of mRFP. N = amino terminus; C = carboxyl terminus.]] | + | [[Image:Haynes_mCherry_PDB2H5Q.png|200px|thumb|mCherry is a mutant derivative of mRFP. PDB 2H5Q is shown here. N = amino terminus; C = carboxyl terminus.]] |
* Protein Data Bank ID 2H5Q | * Protein Data Bank ID 2H5Q | ||
| + | <br> | ||
"Monomeric cherry" (mCherry) red fluorescent protein is a derivative of monomeric RFP from the mushroom coral ''Discosoma sp.'' (Shaner et al. 2004, Shu et al. 2006). This BioBrick was codon optimized (by the lab of Pam Silver) for expression in human/ mammalian cells. Only a single copy is needed for detectable signal. We have assembled two copies in tandem to create a "bulky" protein for use as an artificial transcriptional repressor (Haynes et al. 2012). | "Monomeric cherry" (mCherry) red fluorescent protein is a derivative of monomeric RFP from the mushroom coral ''Discosoma sp.'' (Shaner et al. 2004, Shu et al. 2006). This BioBrick was codon optimized (by the lab of Pam Silver) for expression in human/ mammalian cells. Only a single copy is needed for detectable signal. We have assembled two copies in tandem to create a "bulky" protein for use as an artificial transcriptional repressor (Haynes et al. 2012). | ||
| − | + | <br> | |
REFERENCES | REFERENCES | ||
| Line 20: | Line 21: | ||
# Haynes KA, Ceroni F, Flicker D, Younger A, Silver PA. (2012) A sensitive switch for visualizing natural gene silencing in single cells. ACS Syn Biol. | # Haynes KA, Ceroni F, Flicker D, Younger A, Silver PA. (2012) A sensitive switch for visualizing natural gene silencing in single cells. ACS Syn Biol. | ||
| − | <br><br><br><br><br><br><br> | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
| + | |||
| + | <h2> <b> NYMU-Taipei 2019 - Characterization </b> </h2> | ||
| + | We were searching for fluorescent protein-coding genes to be used in our project. This BBa_J176005 iGEM part was listed in the well 1C of Plate 6 of the 2018 Spring iGEM Distribution Kit. The plasmid backbone listed was pSB1C3 (see https://parts.igem.org/partsdb/get_part.cgi?part=BBa_J176005). We had tried to do transformation with the provided plasmid DNA on E. coli DH5_alpha cells and grow the transformed cells on Chloramphenicol plates. However, no mCherry red color had shown up on the plates. Therefore, we would like to let other iGEM teams know that this BBa_J176005 contains just a coding sequence with no promoter to be expressed in E. coli. | ||
| + | <br> | ||
| + | <br> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_J176005 SequenceAndFeatures</partinfo> | <partinfo>BBa_J176005 SequenceAndFeatures</partinfo> | ||
Latest revision as of 15:33, 16 October 2019
mCh
Codon optimized mCherry red fluorescent protein
Usage and Biology
- Protein Data Bank ID 2H5Q
"Monomeric cherry" (mCherry) red fluorescent protein is a derivative of monomeric RFP from the mushroom coral Discosoma sp. (Shaner et al. 2004, Shu et al. 2006). This BioBrick was codon optimized (by the lab of Pam Silver) for expression in human/ mammalian cells. Only a single copy is needed for detectable signal. We have assembled two copies in tandem to create a "bulky" protein for use as an artificial transcriptional repressor (Haynes et al. 2012).
REFERENCES
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004) Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 22:1567-1572
- Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ (2006) Novel chromophores and buried charges control color in mFruits. Biochem. 45:9639-9647.
- Haynes KA, Ceroni F, Flicker D, Younger A, Silver PA. (2012) A sensitive switch for visualizing natural gene silencing in single cells. ACS Syn Biol.
NYMU-Taipei 2019 - Characterization
We were searching for fluorescent protein-coding genes to be used in our project. This BBa_J176005 iGEM part was listed in the well 1C of Plate 6 of the 2018 Spring iGEM Distribution Kit. The plasmid backbone listed was pSB1C3 (see https://parts.igem.org/partsdb/get_part.cgi?part=BBa_J176005). We had tried to do transformation with the provided plasmid DNA on E. coli DH5_alpha cells and grow the transformed cells on Chloramphenicol plates. However, no mCherry red color had shown up on the plates. Therefore, we would like to let other iGEM teams know that this BBa_J176005 contains just a coding sequence with no promoter to be expressed in E. coli.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]