Difference between revisions of "Part:BBa K592008"
| (3 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
T5-lac promoter is a hybrid promoter made from the phage T5 early promoter and the lac-operon. It contains three LacI binding sites and remains repressed in LacIq strains where LacI is expressed in high levels. It is inducible by IPTG, and recognizable by ''E coli'' RNA polymerase. Compared to the T7 promoters, T5-lac promoter doesn't require the co-expression of phage polymerase. | T5-lac promoter is a hybrid promoter made from the phage T5 early promoter and the lac-operon. It contains three LacI binding sites and remains repressed in LacIq strains where LacI is expressed in high levels. It is inducible by IPTG, and recognizable by ''E coli'' RNA polymerase. Compared to the T7 promoters, T5-lac promoter doesn't require the co-expression of phage polymerase. | ||
| − | + | ===Usage and Biology=== | |
| − | + | ||
| − | + | ||
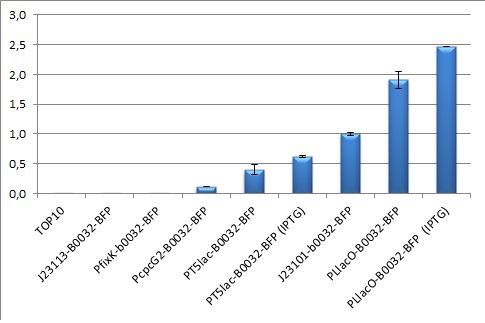
[[Image:Promoter characterization.png]] | [[Image:Promoter characterization.png]] | ||
| + | '''iGEM11_Uppsala-Sweden:''' Promoter characterization. We characterised the promotor in a TOP10 ''E. coli'' strain together with PcpcG2 (<partinfo>BBa_K592003</partinfo>), PLlacO (<partinfo>BBa_R0011</partinfo>), <partinfo>BBa_J23113</partinfo>, PfixK (<partinfo>BBa_K592006</partinfo>) and as reference promoter we used <partinfo>J23101</partinfo>. The backbone pSB3K3, the RBS <partinfo>BBa_B0032</partinfo> and the reporter BFP (<partinfo>BBa_K592100</partinfo>) were used in every construct, and we calculated the relative promoter units, RPU, compared to J23101. The actual test was done in a slightly different way than that presented at the parts.igem. Here we grew the cells in LB medium for 10 hours and we used flow cytometry to quantify the output. The flow cytometer used was a BD FACSAria II, using a 407 nm violet laser and a DAPI filter (band pass 450/40). | ||
| + | |||
| + | The results showed that PT5lac has a strength of 0.4 RPU in TOP10 when repressed by LacI (not lacIq) and a strength of 0.6 RPU when induced with IPTG. | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/8045263] Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Muller-Hill B (1994) Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J 13: 3348-3355. | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/3900050] Gentz R, Bujard H (1985) Promoters Recognized by Escherichia-Coli Rna-Polymerase Selected by Function - Highly Efficient Promoters from Bacteriophage-T5. J Bacteriol 164: 70-77. | ||
| + | |||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/3057497] Lanzer M, Bujard H (1988) Promoters Largely Determine the Efficiency of Repressor Action. P Natl Acad Sci USA 85: 8973-8977. | ||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
Latest revision as of 13:49, 22 September 2011
T5-lac Promoter
T5-lac promoter is a hybrid promoter made from the phage T5 early promoter and the lac-operon. It contains three LacI binding sites and remains repressed in LacIq strains where LacI is expressed in high levels. It is inducible by IPTG, and recognizable by E coli RNA polymerase. Compared to the T7 promoters, T5-lac promoter doesn't require the co-expression of phage polymerase.
Usage and Biology
iGEM11_Uppsala-Sweden: Promoter characterization. We characterised the promotor in a TOP10 E. coli strain together with PcpcG2 (BBa_K592003), PLlacO (BBa_R0011), BBa_J23113, PfixK (BBa_K592006) and as reference promoter we used BBa_J23101. The backbone pSB3K3, the RBS BBa_B0032 and the reporter BFP (BBa_K592100) were used in every construct, and we calculated the relative promoter units, RPU, compared to J23101. The actual test was done in a slightly different way than that presented at the parts.igem. Here we grew the cells in LB medium for 10 hours and we used flow cytometry to quantify the output. The flow cytometer used was a BD FACSAria II, using a 407 nm violet laser and a DAPI filter (band pass 450/40).
The results showed that PT5lac has a strength of 0.4 RPU in TOP10 when repressed by LacI (not lacIq) and a strength of 0.6 RPU when induced with IPTG.
References
[http://www.ncbi.nlm.nih.gov/pubmed/8045263] Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Muller-Hill B (1994) Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J 13: 3348-3355.
[http://www.ncbi.nlm.nih.gov/pubmed/3900050] Gentz R, Bujard H (1985) Promoters Recognized by Escherichia-Coli Rna-Polymerase Selected by Function - Highly Efficient Promoters from Bacteriophage-T5. J Bacteriol 164: 70-77.
[http://www.ncbi.nlm.nih.gov/pubmed/3057497] Lanzer M, Bujard H (1988) Promoters Largely Determine the Efficiency of Repressor Action. P Natl Acad Sci USA 85: 8973-8977.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]