Difference between revisions of "Part:BBa K382021"
| (8 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
Miraculin is a 'flavor inverting' protein, found naturally in the fruit of the plant fruit of Synsepalum dulcificum. Not sweet by itself, miraculin binds to taste receptors on the tongue, possibly altering the structure of the receptors and causing traditionally 'sour' flavors to be received as 'sweet'. | Miraculin is a 'flavor inverting' protein, found naturally in the fruit of the plant fruit of Synsepalum dulcificum. Not sweet by itself, miraculin binds to taste receptors on the tongue, possibly altering the structure of the receptors and causing traditionally 'sour' flavors to be received as 'sweet'. | ||
| − | BBa_K382021 is an Arabidopsis codon-optimized version of miraculin. We characterized expression of YFP-tagged versions in ''E. coli''. The C-terminal 2xYFP tagged version has been submitted to the registry ([[Part:BBa_K382040|BBa_K382040]]). | + | BBa_K382021 is an ''Arabidopsis'' codon-optimized version of miraculin. We characterized expression of YFP-tagged versions in ''E. coli''. The C-terminal 2xYFP tagged version has been submitted to the registry ([[Part:BBa_K382040|BBa_K382040]]). 2xYFP-tagged miraculin was expressed from a IPTG-inducible Novagen Duet vector that was adapted to the BioBrick standard (described in [http://www.jbioleng.org/content/4/1/3 Agapakis et al., JBE, 2010]).<br> |
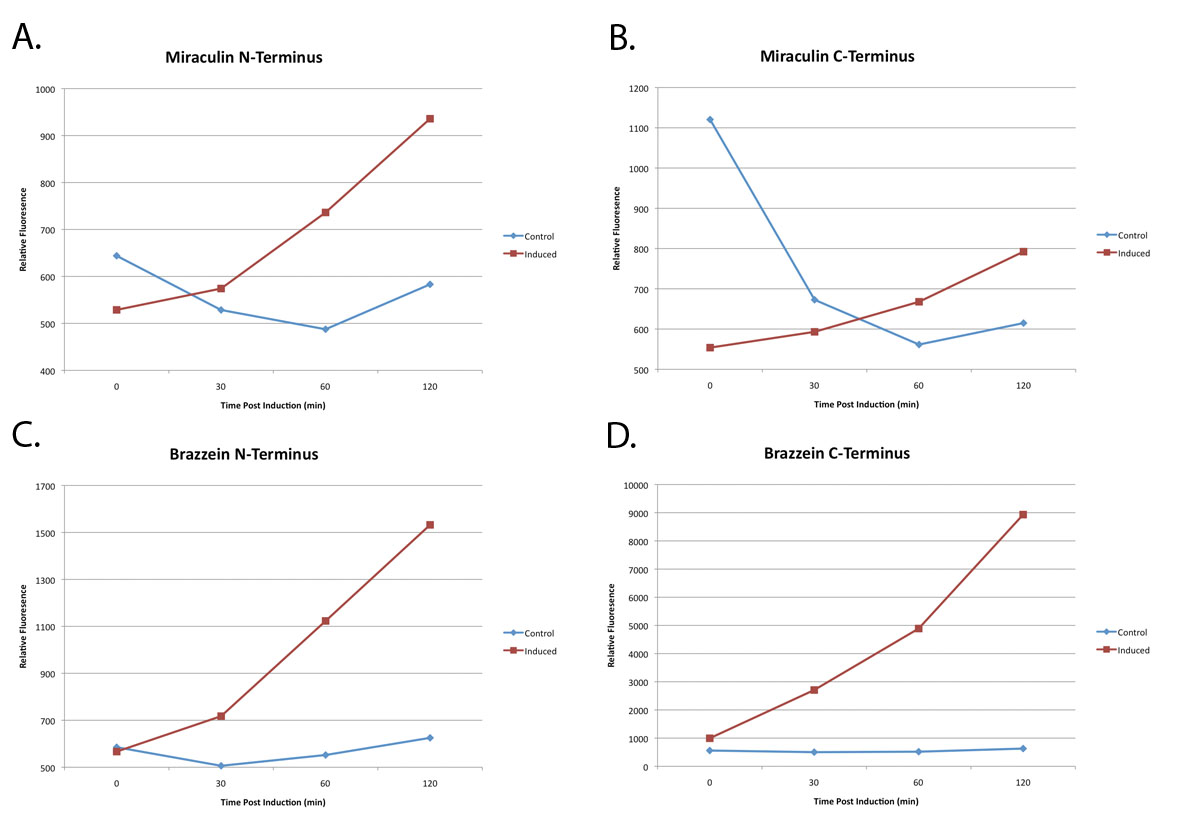
| − | The graphs below show IPTG-induction of miraculin over time in E. coli liquid cell culture (red line) compared to uninduced cultures (blue line). 2xYFP tags were attached to either N- (panel A) or C- terminus (panel B) of miraculin to ensure that YFP position had no influence over expression. In | + | The graphs below show IPTG-induction of miraculin over time in ''E. coli'' liquid cell culture (red line) compared to uninduced cultures (blue line). 2xYFP tags were attached to either N- (panel A) or C- terminus (panel B) of miraculin to ensure that YFP position had no influence over expression. In both cases relative miraculin expression (indicated by YFP fluorescence) had appreciably increased after 120 minutes as compared to the non-induced ''E. Coli''.<br> |
| + | Note: Panels (C) and (D) show similar characterization of a different flavor protein, [[Part:BBa_K382020|Arabidopsis codon-optimized brazzein, BBa_K382020]]. | ||
<br> | <br> | ||
| + | |||
[[Image:Mir_BrazzYFP_Fig_1-crop.jpeg|600px]] | [[Image:Mir_BrazzYFP_Fig_1-crop.jpeg|600px]] | ||
| − | |||
| − | We <br> | + | |
| + | '''Confirmed function''': We have confirmed that the BioBricked ''Arabidopsis'' codon-optimized miraculin protein can be tagged with YFP and expressed from an IPTG inducible promoter in ''E. coli''. Due to safety restrictions, the flavor inverting function of miraculin expressed from ''E. coli'' has not been tested. Continued work will allow us to produce miraculin in a form that is safe for taste testing.<br> | ||
| + | |||
| + | If you are considering producing and testing miraculin yourself, <b>please consult your local bio-hazard/ bio-safety organization for guidelines on producing miraculin that is safe to consume.</b> | ||
Latest revision as of 18:06, 5 November 2010
Arabidopsis optimized Miraculin
Miraculin is a 'flavor inverting' protein, found naturally in the fruit of the plant fruit of Synsepalum dulcificum. Not sweet by itself, miraculin binds to taste receptors on the tongue, possibly altering the structure of the receptors and causing traditionally 'sour' flavors to be received as 'sweet'.
BBa_K382021 is an Arabidopsis codon-optimized version of miraculin. We characterized expression of YFP-tagged versions in E. coli. The C-terminal 2xYFP tagged version has been submitted to the registry (BBa_K382040). 2xYFP-tagged miraculin was expressed from a IPTG-inducible Novagen Duet vector that was adapted to the BioBrick standard (described in [http://www.jbioleng.org/content/4/1/3 Agapakis et al., JBE, 2010]).
The graphs below show IPTG-induction of miraculin over time in E. coli liquid cell culture (red line) compared to uninduced cultures (blue line). 2xYFP tags were attached to either N- (panel A) or C- terminus (panel B) of miraculin to ensure that YFP position had no influence over expression. In both cases relative miraculin expression (indicated by YFP fluorescence) had appreciably increased after 120 minutes as compared to the non-induced E. Coli.
Note: Panels (C) and (D) show similar characterization of a different flavor protein, Arabidopsis codon-optimized brazzein, BBa_K382020.
Confirmed function: We have confirmed that the BioBricked Arabidopsis codon-optimized miraculin protein can be tagged with YFP and expressed from an IPTG inducible promoter in E. coli. Due to safety restrictions, the flavor inverting function of miraculin expressed from E. coli has not been tested. Continued work will allow us to produce miraculin in a form that is safe for taste testing.
If you are considering producing and testing miraculin yourself, please consult your local bio-hazard/ bio-safety organization for guidelines on producing miraculin that is safe to consume.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]