Difference between revisions of "Part:BBa K404122"
| Line 7: | Line 7: | ||
<html> | <html> | ||
| − | + | <p class="MsoTocHeading">Contents</p> | |
| − | + | ||
| − | <p class=MsoTocHeading>Contents</p> | + | |
| − | <p class=MsoToc2 | + | <p class="MsoToc2"><a href="#_Toc275981841"><span lang="EN-US">Arming: Suicide |
| − | lang=EN-US>Arming: Suicide Genes as GOIs</span><span style= | + | Genes as GOIs</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">1</span></a></p> |
| − | display:none;text-decoration:none | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981842"><span lang="EN-US">Introduction</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">1</span></a></p> |
| − | lang=EN-US>Introduction</span><span style= | + | |
| − | text-decoration:none | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981843"><span lang="EN-US">Successful Assembly |
| − | lang=EN-US>Successful Assembly of Vector Plasmids Carrying Suicide Genes via | + | of Vector Plasmids Carrying Suicide Genes via Cloning</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">1</span></a></p> |
| − | Cloning</span><span style= | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981844"><span lang="EN-US">Monitoring Efficient |
| − | lang=EN-US>Monitoring Efficient Tumor Killing by Phase-Contrast Microscopy</span><span | + | Tumor Killing by Phase-Contrast Microscopy</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">4</span></a></p> |
| − | style= | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981845"><span lang="EN-US">Quantitative |
| − | lang=EN-US>Quantitative Analysis of Cell Death by Flow Cytometry</span><span | + | Analysis of Cell Death by Flow Cytometry</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">6</span></a></p> |
| − | style= | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981846"><span lang="EN-US">Titrating |
| − | lang=EN-US>Titrating Ganciclovir Concentrations for Efficient Cell Killing by | + | Ganciclovir Concentrations for Efficient Cell Killing by Cytotoxicity Assays</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">8</span></a></p> |
| − | Cytotoxicity Assays</span><span style= | + | |
| − | text-decoration:none | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981847"><span lang="EN-US">Killing Untransduced |
| − | lang=EN-US>Killing Untransduced Tumor Cells via Bystander Effect</span><span | + | Tumor Cells via Bystander Effect</span><span style="color: windowtext; display: none; text-decoration: none;"> </span><span style="color: windowtext; display: none; text-decoration: none;">13</span></a></p> |
| − | style= | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981848"><span lang="EN-US">Conclusions</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">16</span></a></p> |
| − | lang=EN-US>Conclusions</span><span style= | + | |
| − | text-decoration:none | + | |
| − | style= | + | |
| − | <p class=MsoToc3 | + | <p class="MsoToc3"><a href="#_Toc275981849"><span lang="EN-US">References</span><span style="color: windowtext; display: none; text-decoration: none;">. </span><span style="color: windowtext; display: none; text-decoration: none;">16</span></a></p> |
| − | lang=EN-US>References</span><span style= | + | |
| − | text-decoration:none | + | |
| − | style= | + | |
| − | <p class=MsoNormal> </p> | + | <p class="MsoNormal"> </p> |
| − | <h2><a name="_Toc275981841"><span lang=EN-US>Arming: Suicide Genes as GOIs</span></a></h2> | + | <h2><a name="_Toc275981841"><span lang="EN-US">Arming: Suicide Genes as GOIs</span></a></h2> |
| − | <h3><a name="_Toc275981842"><span lang=EN-US>Introduction</span></a></h3> | + | <h3><a name="_Toc275981842"><span lang="EN-US">Introduction</span></a></h3> |
| − | <p class=MsoNormal><span lang=EN-US>Gene delivery using viral vectors to | + | <p class="MsoNormal"><span lang="EN-US">Gene delivery using viral vectors to |
specifically target tumor cells gained increasing attention in the last years | specifically target tumor cells gained increasing attention in the last years | ||
| − | being efficient in combination with suicide gene approaches </span><span lang=EN-US>(Willmon et al. 2006)</span><span lang=EN-US>. Several prodrug/enzyme | + | being efficient in combination with suicide gene approaches </span><span lang="EN-US">(Willmon et al. 2006)</span><span lang="EN-US">. Several prodrug/enzyme |
| − | combinations have been reported. Two systems - ganciclovir (GCV)/herpes | + | combinations have been reported. Two systems - ganciclovir (GCV)/herpes simplex |
| − | simplex virus thymidine kinase (HSV-TK) </span><span | + | virus thymidine kinase (HSV-TK) </span><span lang="EN-US">(Ardiani et al. 2010)</span><span lang="EN-US"> and |
| − | lang=EN-US>(Ardiani et al. 2010)</span><span lang=EN-US> and | + | 5-fluorocytosine/cytosine deaminase (CD) </span><span lang="EN-US">(Fuchita et al. 2009a)</span><span lang="EN-US"> – have been widely |
| − | 5-fluorocytosine/cytosine deaminase (CD) </span><span | + | used and their therapeutic benefit was demonstrated in preclinical studies </span><span lang="EN-US">(Greco & Dachs 2001)</span><span lang="EN-US">. Adeno-associated |
| − | lang=EN-US>(Fuchita et al. 2009a)</span><span lang=EN-US> – have been widely | + | |
| − | used and their therapeutic benefit was demonstrated in preclinical studies </span><span lang=EN-US>(Greco & Dachs 2001)</span><span lang=EN-US>. Adeno-associated | + | |
viruses (AAV) as delivery vectors are commonly used in suicide gene therapy. The | viruses (AAV) as delivery vectors are commonly used in suicide gene therapy. The | ||
| − | suicide gene flanked by the inverted terminal repeats (ITRs) is encapsulated | + | suicide gene flanked by the inverted terminal repeats (ITRs) is encapsulated into |
| − | into the virus particles and delivered to the target cells where suicide gene | + | the virus particles and delivered to the target cells where suicide gene |
expression is mediated by cellular proteins.</span></p> | expression is mediated by cellular proteins.</span></p> | ||
| − | <p class=MsoNormal><span lang=EN-US>The iGEM team Freiburg_Bioware 2010 provides | + | <p class="MsoNormal"><span lang="EN-US">The iGEM team Freiburg_Bioware 2010 provides |
| − | both the cytosine deaminase (CD, BBa_K404112) and an improved guanylate kinase | + | both the cytosine deaminase (CD, BBa_K404112) and an improved guanylate kinase - |
| − | - thymidine kinase fusion gene (mGMK_TK, BBa_K404113) within the Virus | + | thymidine kinase fusion gene (mGMK_TK, BBa_K404113) within the Virus |
Construction Kit as effective suicide genes. We demonstrate efficient and | Construction Kit as effective suicide genes. We demonstrate efficient and | ||
specific killing of tumor cells by enzymatic cytotoxicity assays, flow | specific killing of tumor cells by enzymatic cytotoxicity assays, flow | ||
| Line 86: | Line 60: | ||
into the viral capsids. </span></p> | into the viral capsids. </span></p> | ||
| − | <h3><a name="_Toc275981843"><span lang=EN-US>Successful Assembly of Vector Plasmids | + | <h3><a name="_Toc275981843"><span lang="EN-US">Successful Assembly of Vector Plasmids |
Carrying Suicide Genes via Cloning</span></a></h3> | Carrying Suicide Genes via Cloning</span></a></h3> | ||
| − | <p class=MsoNormal><span lang=EN-US> | + | <p class="MsoNormal"><span lang="EN-US">To create the functional vector plasmids, assembly |
| − | suicide genes | + | of the constructs carrying the suicide genes was performed following the |
| − | Standard Assembly. All plasmids contain the enhancer-element human <i>beta-globin</i> | + | BioBrick Standard Assembly. All plasmids contain the enhancer-element human <i>beta-globin</i> |
intron (BBa_K404107) and the <i>human growth hormone</i> terminator signal (hGH, | intron (BBa_K404107) and the <i>human growth hormone</i> terminator signal (hGH, | ||
BBa_K404108) flanked by the inverted terminal repeats (ITRs, BBa_K404100 and | BBa_K404108) flanked by the inverted terminal repeats (ITRs, BBa_K404100 and | ||
BBa_K404101). Assembled suicide genes are either under the control of the CMV | BBa_K404101). Assembled suicide genes are either under the control of the CMV | ||
| − | promoter or the tumor-specific telomerase promoter phTERT (BBa_K404106). </span></p> | + | promoter (BBa_J52034) or the tumor-specific telomerase promoter phTERT (BBa_K404106). |
| + | </span></p> | ||
| − | < | + | <div align="center"> |
| − | + | <table class="MsoTableGrid" style="border: medium none ; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0"> | |
| − | + | <tbody><tr style="height: 343.9pt;"> | |
| − | + | <td style="padding: 0cm 5.4pt; width: 435.8pt; height: 343.9pt;" valign="top" width="581"> | |
| − | + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 581px; height: 394px;" alt="" id="Grafik 9" src="https://static.igem.org/mediawiki/2010/9/9b/Freiburg10_BBA_overview_suicide.png"></p> | |
| − | <table class=MsoTableGrid | + | <p class="MsoCaption"><a name="_Ref275976895"><span lang="EN-US">Figure </span></a><span lang="EN-US">1</span><span lang="EN-US">: BioBrick-compatible assembly of functional |
| − | + | ||
| − | <tr style= | + | |
| − | <td | + | |
| − | + | ||
| − | <p class=MsoNormal style= | + | |
| − | + | ||
| − | + | ||
| − | <p class=MsoCaption><a name="_Ref275976895"><span lang=EN-US>Figure </span></a><span lang=EN-US>1</span><span lang=EN-US>: BioBrick-compatible assembly of functional | + | |
vector plasmids containing the suicide genes. The schematic figure shows the | vector plasmids containing the suicide genes. The schematic figure shows the | ||
cloning strategy of the guanylate kinase – thymidine kinase fusion gene | cloning strategy of the guanylate kinase – thymidine kinase fusion gene | ||
| Line 117: | Line 84: | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span lang="EN-US"> </span></p> |
| − | <p class=MsoNormal><span lang=EN-US> | + | <p class="MsoNormal"><span lang="EN-US">To enable modularization of the thymidine |
| − | mutants TK30 and SR39 (BBa_K404109 and BBa_K404110) according to the | + | kinase mutants TK30 and SR39 (BBa_K404109 and BBa_K404110) according to the |
| − | standard, the fusion genes mGMK_TK30 and mGMK_SR39 (BBa_K404113 and </span><span | + | BioBrick standard, the fusion genes mGMK_TK30 and mGMK_SR39 (BBa_K404113 and </span><span style="line-height: 200%; color: black;" lang="EN-US">BBa_K404315</span><span lang="EN-US">) and CD (</span><span style="line-height: 200%; color: black;" lang="EN-US">BBa_K404112</span><span lang="EN-US">) were modified using the QuikChange |
| − | + | Lightning Site-Directed Mutagenesis Kit (Stratagene) for deletion of iGEM RFC10 | |
| − | lang=EN-US>) and CD (</span><span | + | pre- and suffix restriction sites. </span><span lang="EN-US">Figure 1</span><span lang="EN-US"> demonstrates one example of |
| − | color:black | + | successful deletion of a PstI restriction site located within the mGMK_TK30 |
| − | Lightning Site-Directed Mutagenesis Kit (Stratagene) for deletion of iGEM pre- | + | sequence at position 3109. A point mutation was introduced replacing the |
| − | and suffix restriction sites. </span><span lang=EN-US>Figure 1</span><span | + | nucleotide G by A, resulting in the deletion of the restriction site while |
| − | lang=EN-US> demonstrates one example of successful deletion of | + | maintaining the encoded amino acid glutamine. Exchange of guanine to adenine |
| − | restriction site located within the mGMK_TK30 sequence at position 3109. | + | was confirmed by sequencing (</span><span lang="EN-US">Figure 2</span><span lang="EN-US">). </span></p> |
| − | + | ||
| − | the deletion of the restriction site | + | |
| − | + | ||
| − | <div align=center> | + | <div align="center"> |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; width: 479.55pt; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0" width="639"> |
| − | + | <tbody><tr style="height: 179.55pt;"> | |
| − | <tr style= | + | <td style="padding: 0cm 5.4pt; width: 479.55pt; height: 179.55pt;" valign="top" width="639"> |
| − | <td | + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 635px; height: 328px;" alt="" id="Grafik 81" src="https://static.igem.org/mediawiki/2010/3/32/Freiburg10_SDM_mGMK_TK30.png"></p> |
| − | + | <p class="MsoCaption"><a name="_Ref275976917"><span lang="EN-US">Figure </span></a><span lang="EN-US">2</span><span lang="EN-US">: Cytosine to guanine exchange by | |
| − | <p class=MsoNormal style= | + | |
| − | + | ||
| − | + | ||
| − | <p class=MsoCaption><a name="_Ref275976917"><span lang=EN-US>Figure </span></a><span lang=EN-US>2</span><span lang=EN-US>: | + | |
site-directed mutagenesis using QuikChange Lightning Kit provided by | site-directed mutagenesis using QuikChange Lightning Kit provided by | ||
| − | Stratagene | + | Stratagene was successful as demonstrated by (A) test digestion linearizing |
| − | + | the plasmid with PstI and (B) by sequencing.</span></p> | |
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span lang="EN-US"> </span></p> |
| − | <p class=MsoNormal><span lang=EN-US>Furthermore, assembly of BioBrick-compatible | + | <p class="MsoNormal"><span lang="EN-US">Furthermore, assembly of BioBrick-compatible |
vector plasmids was performed. An example for the last assembly step of | vector plasmids was performed. An example for the last assembly step of | ||
| − | mGMK_TK30 and hGH_rITR is shown in </span><span | + | mGMK_TK30 and hGH_rITR is shown in </span><span lang="EN-US">Figure 3</span><span lang="EN-US">. The plasmids were digested with |
| − | lang=EN-US>Figure 3</span><span lang=EN-US>. The plasmids were digested with | + | both XbaI and PstI (New England Biolabs, Insert: BBa_K404116: hGH_rITR) or SpeI |
| − | both XbaI and PstI (Insert: BBa_K404116: hGH_rITR) or SpeI and PstI (Vector) and | + | and PstI (Vector) and loaded on an agarose gel. As demonstrated in the preparative |
| − | loaded on an agarose gel. As demonstrated in the preparative gel in </span><span lang=EN-US>Figure 3</span><span lang=EN-US>, the expected bands were detected | + | gel in </span><span lang="EN-US">Figure 3</span><span lang="EN-US">, the expected |
| − | under UV light and the extracted DNA was | + | bands were detected under UV light and the extracted, subsequently purified DNA |
| − | step for producing BioBricks was conducted following the iGEM BioBrick | + | was successfully ligated and transformed into <i>E. coli</i>. Each assembly |
| + | step for producing the BioBricks was conducted following the iGEM BioBrick | ||
standard.</span></p> | standard.</span></p> | ||
| − | <div align=center> | + | <div align="center"> |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; width: 387.35pt; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0" width="516"> |
| − | + | <tbody><tr style="height: 125.95pt;"> | |
| − | <tr style= | + | <td style="padding: 0cm 5.4pt; width: 387.35pt; height: 125.95pt;" valign="top" width="516"> |
| − | <td | + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 519px; height: 189px;" alt="" id="Grafik 28" src="https://static.igem.org/mediawiki/2010/f/f2/Freiburg10_Cloning_mGMK_TK30_Arming.png"></p> |
| − | + | <p class="MsoCaption"><a name="_Ref275976959"><span lang="EN-US">Figure </span></a><span lang="EN-US">3</span><span lang="EN-US">: Assembly of mGMK_TK30 (vector molecule, in | |
| − | <p class=MsoNormal style= | + | pSB1C3) and hGH-terminator_rightITR (insert molecule). The digested fragments |
| − | + | were visualized under UV-light and correspond to the expected sizes.</span></p> | |
| − | + | ||
| − | <p class=MsoCaption><a name="_Ref275976959"><span lang=EN-US>Figure </span></a><span lang=EN-US>3</span><span lang=EN-US>: | + | |
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span lang="EN-US"> </span></p> |
| − | <h3><a name="_Toc275981844"><span lang=EN-US>Monitoring Efficient Tumor Killing | + | <h3><a name="_Toc275981844"><span lang="EN-US">Monitoring Efficient Tumor Killing |
by Phase-Contrast Microscopy</span></a></h3> | by Phase-Contrast Microscopy</span></a></h3> | ||
| − | <p class=MsoNormal><span lang=EN-US>Tumor cells, transduced with viral | + | <p class="MsoNormal"><span lang="EN-US">Tumor cells, transduced with viral |
particles encapsidating the effector constructs containing the mGMK_TK30 driven | particles encapsidating the effector constructs containing the mGMK_TK30 driven | ||
| − | by the CMV promoter, were cultured in presence and absence of ganciclovir. | + | by the CMV promoter, were cultured both in presence and absence of ganciclovir |
| − | Morphological changes were monitored via phase-contrast microscopy | + | (Roche). Morphological changes were monitored via phase-contrast microscopy for |
| − | hours post infection. As it can be seen in </span><span | + | 48 hours post infection. As it can be seen in </span><span lang="EN-US">Figure 4</span><span lang="EN-US">, non-transduced tumor cells treated |
| − | lang=EN-US>Figure 4</span><span lang=EN-US> non-transduced cells treated with ganciclovir | + | with ganciclovir and transduced cells without ganciclovir did not show |
| − | and transduced | + | significant cell ablation. In contrast, transduced cells expressing the |
| − | ablation. In contrast transduced cells expressing the guanylate kinase - | + | guanylate kinase - thymidine kinase fusion protein showed significant cell |
| − | thymidine kinase fusion protein | + | death after incubation with ganciclovir for 48 hours post infection. </span></p> |
| − | with ganciclovir for 48 hours post infection. </span></p> | + | |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; width: 491pt; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0" width="655"> |
| − | + | <tbody><tr style="height: 188.8pt;"> | |
| − | <tr style= | + | <td style="padding: 0cm 5.4pt; width: 247.35pt; height: 188.8pt;" width="330"> |
| − | <td | + | <p class="MsoNoSpacing"><b><span lang="EN-US">A</span></b><span lang="EN-US"> </span><img style="width: 318px; height: 238px;" id="Grafik 1" src="https://static.igem.org/mediawiki/2010/4/4f/Freiburg10_Microscopy_Arming_A.png" alt="Beschreibung: https://static.igem.org/mediawiki/2010/3/3e/HT_negativ_control_ganciclovir_only.jpg"></p> |
| − | <p class=MsoNoSpacing><b><span lang=EN-US>A</span></b><span lang=EN-US> </span><img | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</td> | </td> | ||
| − | <td | + | <td style="padding: 0cm 5.4pt; width: 243.65pt; height: 188.8pt;" width="325"> |
| − | <p class=MsoNoSpacing><b><span lang=EN-US>B</span></b><span lang=EN-US> </span><img | + | <p class="MsoNoSpacing"><b><span lang="EN-US">B</span></b><span lang="EN-US"> </span><img style="width: 318px; height: 238px;" id="Grafik 2" src="https://static.igem.org/mediawiki/2010/e/e8/Freiburg10_Microscopy_Arming_B.png" alt="Beschreibung: https://static.igem.org/mediawiki/2010/f/f0/HT_TKGMK_clone_1_ohne_Ganciclovir_300%C2%B5l.jpg"></p> |
| − | + | ||
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | <tr style= | + | <tr style="height: 188.8pt;"> |
| − | <td | + | <td style="padding: 0cm 5.4pt; width: 247.35pt; height: 188.8pt;" width="330"> |
| − | <p class=MsoNoSpacing><b><span lang=EN-US>C</span></b><span lang=EN-US> </span></p> | + | <p class="MsoNoSpacing"><b><span lang="EN-US">C</span></b><span lang="EN-US"> </span></p> |
| − | <p class=MsoNoSpacing style= | + | <p class="MsoNoSpacing" style="page-break-after: avoid;"><img style="width: 318px; height: 239px;" id="Grafik 3" src="https://static.igem.org/mediawiki/2010/a/ac/Freiburg10_Microscopy_Arming_C.png" alt="Beschreibung: https://static.igem.org/mediawiki/2010/c/c3/HT_TKGMK_clone_1_with_ganciclovir_300%C2%B5l.jpg"></p> |
| − | + | ||
| − | + | ||
| − | + | ||
</td> | </td> | ||
| − | <td | + | <td style="padding: 0cm 5.4pt; width: 243.65pt; height: 188.8pt;" width="325"> |
| − | <p class=MsoNoSpacing><b><span lang=EN-US>D</span></b><span lang=EN-US> </span><img | + | <p class="MsoNoSpacing"><b><span lang="EN-US">D</span></b><span lang="EN-US"> </span><img style="width: 318px; height: 238px;" id="Grafik 4" src="https://static.igem.org/mediawiki/2010/d/df/Freiburg10_Microscopy_Arming_D.png" alt="Beschreibung: https://static.igem.org/mediawiki/2010/7/73/HT_TKGMK_clone_1_with_ganciclovir_600%C2%B5l_well_2.jpg"></p> |
| − | + | ||
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | <tr style= | + | <tr style="height: 34.2pt;"> |
| − | <td | + | <td colspan="2" style="padding: 0cm 5.4pt; width: 491pt; height: 34.2pt;" width="655"> |
| − | + | <p class="MsoCaption"><a name="_Ref275976998"><span lang="EN-US">Figure </span></a><span lang="EN-US">4</span><span lang="EN-US">: Qualitative analysis of cell death induced | |
| − | <p class=MsoCaption><a name="_Ref275976998"><span lang=EN-US>Figure </span></a><span lang=EN-US>4</span><span lang=EN-US>: Qualitative analysis of cell death induced | + | |
by conversion of ganciclovir to ganciclovir-triphosphate by virus-delivered | by conversion of ganciclovir to ganciclovir-triphosphate by virus-delivered | ||
guanylate - thymidine kinase (mGMK_TK30). A: Non-transduced HT1080 cells | guanylate - thymidine kinase (mGMK_TK30). A: Non-transduced HT1080 cells | ||
| − | incubated in the presence of ganciclovir | + | incubated in the presence of ganciclovir not exhibiting cell death. B: Untreated |
| − | + | transduced HT1080 cells showing a high survival rate. C: HT1080 cells | |
| − | + | transduced with 300 µL viral particles and incubated with ganciclovir leading | |
| − | + | to tumor cell ablation. D: HT1080 cells transduced with 600 µL viral | |
| − | + | particles and incubated with ganciclovir leading to ablation of tumor cells. </span></p> | |
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
| − | <p class=MsoNormal><span lang=EN-US> </span></p> | + | <p class="MsoNormal"><span lang="EN-US"> </span></p> |
| − | <p class=MsoNormal><span lang=EN-US>Suicide gene therapy is based on the | + | <p class="MsoNormal"><span lang="EN-US">Suicide gene therapy is based on the localized |
| − | conversion of non-toxic prodrugs to toxic substances </span><span | + | conversion of non-toxic prodrugs to toxic substances </span><span lang="EN-US">(Greco & Dachs 2001)</span><span lang="EN-US">, promoting cell |
| − | lang=EN-US>(Greco & Dachs 2001)</span><span lang=EN-US>, | + | death in the tumor tissue (</span><span lang="EN-US">Figure 5</span><span lang="EN-US">). Directed gene delivery is achieved by using recombinant viral |
| − | death (</span><span lang=EN-US>Figure 5</span><span lang=EN-US>). Directed gene | + | vectors as provided by the iGEM team Freiburg_Bioware 2010 within the Virus |
| − | delivery is achieved by using recombinant viral vectors as provided by the iGEM | + | Construction Kit.</span></p> |
| − | team Freiburg_Bioware 2010 within the Virus Construction Kit.</span></p> | + | |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; border-collapse: collapse; margin-left: 4.8pt; margin-right: 4.8pt;" align="left" border="0" cellpadding="0" cellspacing="0"> |
| − | + | <tbody><tr style="height: 174.85pt;"> | |
| − | + | <td style="padding: 0cm 5.4pt; width: 447.8pt; height: 174.85pt;" valign="top" width="597"> | |
| − | <tr style= | + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><span style="position: relative; z-index: 251659264; left: -1px; top: 0px; width: 583px; height: 271px;"><img style="width: 597px; height: 279px;" alt="" src="https://static.igem.org/mediawiki/2010/a/a4/Freiburg10_overview_GDEPT.png"></span><br clear="all"> |
| − | <td | + | |
| − | + | ||
| − | <p class=MsoNormal style= | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
</p> | </p> | ||
| − | <p class=MsoCaption><a name="_Ref275977093"><span lang=EN-US>Figure </span></a><span lang=EN-US>5</span><span lang=EN-US>: Overview | + | <p class="MsoCaption"><a name="_Ref275977093"><span lang="EN-US">Figure </span></a><span lang="EN-US">5</span><span lang="EN-US">: Overview of the suicide gene therapy |
approach. Non-toxic prodrugs are converted into toxic effector molecules | approach. Non-toxic prodrugs are converted into toxic effector molecules | ||
leading to cell death of the tumor cells. </span></p> | leading to cell death of the tumor cells. </span></p> | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
| − | <p class=MsoNormal | + | <p class="MsoNormal"><span lang="EN-US">Non-transduced cells can survive in the |
| + | presence of ganciclovir since the prodrug is not toxic for these cells (</span><span lang="EN-US">Figure 4</span><span lang="EN-US">A). </span>The demonstration that | ||
| + | transduced cells are viable in the absence of ganciclovir confirms that cell | ||
| + | killing is indeed induced by combination of the delivered thymidine kinase and | ||
| + | treatment with ganciclovir. Viral particles encapsidating the suicide construct | ||
| + | mGMK_TK30 are efficient in directed gene delivery, thus leading to cell death | ||
| + | of transduced cells due to overexpression of mGMK_TK30 and prodrug conversion. The | ||
| + | cell-toxic ganciclovir-triphosphate is incorporated into the<span lang="EN-US"> | ||
| + | nascent DNA chain leading to replication termination and finally resulting in | ||
| + | death of dividing cells. </span></p> | ||
| − | + | <h3><a name="_Toc275981845"><span lang="EN-US">Quantitative Analysis of Cell | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | <h3><a name="_Toc275981845"><span lang=EN-US>Quantitative Analysis of Cell | + | |
Death by Flow Cytometry</span></a></h3> | Death by Flow Cytometry</span></a></h3> | ||
| − | <p class=MsoNormal><span lang=EN-US>Quantitative analysis of the cytotoxic | + | <p class="MsoNormal"><span lang="EN-US">Quantitative analysis of the cytotoxic |
effect induced by mGMK_TK30 was first conducted by flow cytometry analysis 72 hours | effect induced by mGMK_TK30 was first conducted by flow cytometry analysis 72 hours | ||
| − | post transduction. HT1080 cells were stained with 7-AAD and Annexin V. 7-AAD | + | post transduction. HT1080 cells were stained with 7-AAD and Annexin V. 7-AAD intercalates |
| − | intercalates in double-stranded DNA after penetrating cell membranes of dead | + | in double-stranded DNA after penetrating cell membranes of dead cells, whereas |
| − | cells, whereas Annexin V | + | Annexin V specifically binds phosphatidylserine which is only accessible during |
| − | accessible during apoptosis. </span><span lang=EN-US>Figure 6</span><span | + | apoptosis. </span><span lang="EN-US">Figure 6</span><span lang="EN-US"> demonstrates |
| − | lang=EN-US> demonstrates the relation between cell death and ganciclovir | + | the relation between cell death and ganciclovir concentration.</span></p> |
| − | concentration.</span></p> | + | |
| − | <div align=center> | + | <div align="center"> |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; width: 477.6pt; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0" width="637"> |
| − | + | <tbody><tr style="height: 248.5pt;"> | |
| − | <tr style= | + | <td style="padding: 0cm 5.4pt; width: 241.8pt; height: 248.5pt;" valign="top" width="322"> |
| − | <td | + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 341px; height: 321px;" alt="" id="Grafik 24" src="https://static.igem.org/mediawiki/2010/9/9d/Freiburg10_FACS_A_D.png"></p> |
| − | + | ||
| − | <p class=MsoNormal style= | + | |
| − | + | ||
| − | + | ||
</td> | </td> | ||
| − | <td | + | <td style="padding: 0cm 5.4pt; width: 235.8pt; height: 248.5pt;" valign="top" width="314"> |
| − | + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 300px; height: 326px;" alt="" id="Grafik 36" src="https://static.igem.org/mediawiki/2010/0/0b/Freiburg10_FACS_E_H.png"></p> | |
| − | <p class=MsoNormal style= | + | <p class="MsoCaption"> </p> |
| − | + | ||
| − | + | ||
| − | <p class=MsoCaption> </p> | + | |
</td> | </td> | ||
</tr> | </tr> | ||
| − | <tr style= | + | <tr style="height: 38pt;"> |
| − | <td | + | <td colspan="2" style="padding: 0cm 5.4pt; width: 477.6pt; height: 38pt;" valign="top" width="637"> |
| − | + | <p class="MsoCaption" style="text-indent: 0cm;"><a name="_Ref275977177"><span lang="EN-US">Figure </span></a><span lang="EN-US">6</span><span lang="EN-US">: Flow | |
| − | <p class=MsoCaption style= | + | cytometry data analysis. </span><span lang="EN-US">A: Gating non-transduced |
| − | + | HT1080 cells (control). B: Non-transduced cells without staining plotted | |
| − | + | against 7-AAD. C: Gating non-transduced cells stained with 7-AAD. D: | |
| − | + | Non-transduced, 7-AAD-stained cells plotted against 7-AAD. E: Gating | |
| − | + | transduced cells (GOI: mGMK_TK30) treated with 485 µM ganciclovir. F: Gated, | |
| − | + | Annexin-V stained cells plotted against AnnV-2 Log. G: Gated cells plotted | |
| − | + | against 7-AAD H: Gated, 7-AAD and Annexin-V stained cells plotted against | |
| − | + | 7-AAD and Annexin-V. Gate R19 comprised Annexin-V and 7-AAD positive cells.</span></p> | |
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span lang="EN-US"> </span></p> |
| − | <p class=MsoNormal | + | <p class="MsoNormal" style="text-align: center; text-indent: 0cm;" align="center"><span lang="EN-US"> </span></p> |
| − | lang=EN-US> </span></p> | + | |
| − | <div align=center> | + | <div align="center"> |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0"> |
| − | + | <tbody><tr> | |
| − | <tr> | + | <td style="padding: 0cm 5.4pt; width: 460.6pt;" valign="top" width="614"> |
| − | <td | + | <p class="MsoNormal" style="text-align: center; text-indent: 0cm; page-break-after: avoid;" align="center"><img style="width: 614px; height: 424px;" alt="" id="Diagramm 8" src="https://static.igem.org/mediawiki/2010/2/29/Freiburg10_Diagram_mGMK_effectFACS.png"></p> |
| − | <p class=MsoNormal | + | <p class="MsoCaption"><a name="_Ref275977227"><span lang="EN-US">Figure </span></a><span lang="EN-US">7</span><span lang="EN-US">: Quantification of flow cytometry data |
| − | + | provided in </span><span lang="EN-US">Figure 6</span><span lang="EN-US">. With | |
| − | + | increasing ganciclovir concentration, the survival rate of cells decreases. 60 | |
| − | <p class=MsoCaption><a name="_Ref275977227"><span lang=EN-US>Figure </span></a><span lang=EN-US>7</span><span lang=EN-US>: Quantification of flow cytometry data | + | % of HT1080 cells treated with 4.85 mM ganciclovir show tumor ablation, |
| − | provided in </span><span lang=EN-US>Figure 6</span><span lang=EN-US>. With | + | however even lower amounts of ganciclovir lead to significant cell death.</span></p> |
| − | increasing ganciclovir concentration, the survival rate of cells decreases. 60 | + | |
| − | of HT1080 cells treated with 4 | + | |
| − | even lower amounts of ganciclovir | + | |
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal | + | <p class="MsoNormal" style="text-align: center; text-indent: 0cm;" align="center"><span lang="EN-US"> </span></p> |
| − | lang=EN-US> </span></p> | + | |
| − | <p class=MsoNormal><span lang=EN-US> | + | <p class="MsoNormal"><span lang="EN-US">The effect of different ganciclovir |
| − | concentrations on transduced HT1080 sarcoma cells. | + | concentrations on transduced HT1080 sarcoma cells was investigated. |
| − | performed with recombinant viral particles encapsidating the mGMK_TK30 prodrug | + | Transduction was performed with recombinant viral particles encapsidating the |
| − | gene. 72 hours post infection cells were stained with 7-AAD and Annexin V. As </span><span lang=EN-US>Figure 7</span><span lang=EN-US> shows | + | mGMK_TK30 prodrug gene. 72 hours post infection, cells were stained with 7-AAD |
| − | the | + | and Annexin V. As </span><span lang="EN-US">Figure 7</span><span lang="EN-US"> shows, |
| + | the fraction of killed transduced cells is proportional to the applied ganciclovir | ||
| + | concentration.</span></p> | ||
| − | <h3><a name="_Toc275981846"><span lang=EN-US>Titrating Ganciclovir | + | <h3><a name="_Toc275981846"><span lang="EN-US">Titrating Ganciclovir |
Concentrations for Efficient Cell Killing by Cytotoxicity Assays</span></a></h3> | Concentrations for Efficient Cell Killing by Cytotoxicity Assays</span></a></h3> | ||
| − | <p class=MsoNormal><span lang=EN-US>Further analysis of the cytotoxic effect | + | <p class="MsoNormal"><span lang="EN-US">Further analysis of the cytotoxic effect |
induced by thymidine kinase converting ganciclovir to the toxic anti-metabolite | induced by thymidine kinase converting ganciclovir to the toxic anti-metabolite | ||
has been performed using MTT assays. 3-(4,5-Dimethylthiazol-2-yl)-2,5diphenyltetrazolium | has been performed using MTT assays. 3-(4,5-Dimethylthiazol-2-yl)-2,5diphenyltetrazolium | ||
| − | bromide), also known as MTT, is a yellow tetrazole, which is reduced to purple insoluble | + | bromide), also known as MTT, is a yellow-colored tetrazole, which is reduced to |
| − | formazan in the presence of NADH and NADPH </span><span | + | purple insoluble formazan in the presence of NADH and NADPH </span><span lang="EN-US">(Roche n.d.)</span><span lang="EN-US">. Colorimetric analysis can be |
| − | lang=EN-US>(Roche n.d.)</span><span lang=EN-US>. | + | |
carried out via spectrometry. Different tumor cell lines, HT1080 and A431, were | carried out via spectrometry. Different tumor cell lines, HT1080 and A431, were | ||
transduced with the recombinant viruses carrying the linear DNA construct | transduced with the recombinant viruses carrying the linear DNA construct | ||
| − | coding for mGMK-TK30 regulated by the CMV promoter and treated with ganciclovir. | + | coding for mGMK-TK30 regulated by the CMV promoter and subsequently treated |
| − | 48 and 72 hours post infection cells were incubated with MTT and absorbance of | + | with ganciclovir. 48 and 72 hours post infection, cells were incubated with MTT |
| − | formazan was quantified. </span></p> | + | and fresh DMEM. After cell lysis by DMSO, absorbance of formazan at 570 nm was |
| + | quantified using a Tecan Sunrise plate reader. </span></p> | ||
| − | <div align=center> | + | <div align="center"> |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; width: 451.05pt; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0" width="601"> |
| − | + | <tbody><tr style="height: 176.7pt;"> | |
| − | <tr style= | + | <td style="padding: 0cm 5.4pt; width: 451.05pt; height: 176.7pt;" valign="top" width="601"> |
| − | <td | + | <p class="MsoNormal" style="text-indent: 0cm;"><b><span lang="EN-US">A</span></b><img style="width: 588px; height: 413px;" alt="" id="Diagramm 95" src="https://static.igem.org/mediawiki/2010/8/86/Freiburg10_MTT_72h_2d.png"></p> |
| − | + | ||
| − | <p class=MsoNormal style= | + | |
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | <tr style= | + | <tr style="height: 176.7pt;"> |
| − | <td | + | <td style="padding: 0cm 3.5pt; width: 451.05pt; height: 176.7pt;" valign="top" width="601"> |
| − | + | <p class="MsoNormal" style="text-indent: 0cm;"><b>B</b> </p> | |
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-align: center; text-indent: 0cm; page-break-after: avoid;" align="center"><img style="width: 593px; height: 409px;" alt="" id="Diagramm 10" src="https://static.igem.org/mediawiki/2010/7/76/Freiburg10_MTT_72h_3d.png"></p> |
| − | <p class=MsoNormal | + | <p class="MsoCaption"><a name="_Ref275977309"><span lang="EN-US">Figure </span></a><span lang="EN-US">8</span><span lang="EN-US">: Effect of ganciclovir on HT1080 cell survival |
| − | + | 72 hours post infection as (A) two dimensional plot of survival of cells and | |
| − | + | (B) three-dimensional plot of ganciclovir, virus particles and cell survival.</span></p> | |
| − | <p class=MsoCaption><a name="_Ref275977309"><span lang=EN-US>Figure </span></a><span lang=EN-US>8</span><span lang=EN-US>: Effect of ganciclovir on HT1080 cell | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span style="font-size: 8pt; line-height: 200%;" lang="EN-US"> </span></p> |
| − | + | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span style="font-size: 8pt; line-height: 200%;" lang="EN-US"> </span></p> |
| − | + | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span style="font-size: 8pt; line-height: 200%;" lang="EN-US"> </span></p> |
| − | + | ||
| − | <div align=center> | + | <div align="center"> |
| − | <table class=MsoTableGrid | + | <table class="MsoTableGrid" style="border: medium none ; width: 415pt; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0" width="553"> |
| − | + | <tbody><tr style="height: 176.7pt;"> | |
| − | <tr style= | + | <td style="padding: 0cm 5.4pt; width: 415pt; height: 176.7pt;" valign="top" width="553"> |
| − | <td | + | <p class="MsoNormal" style="text-indent: 0cm;"><b><span lang="EN-US">A </span></b><span lang="EN-US"> </span></p> |
| − | + | <p class="MsoNormal" style="text-align: center; text-indent: 0cm;" align="center"><img style="width: 607px; height: 427px;" alt="" id="Diagramm 1154" src="https://static.igem.org/mediawiki/2010/2/2a/Freiburg10_MTT_96h_2d.png"></p> | |
| − | <p class=MsoNormal style= | + | |
| − | + | ||
| − | <p class=MsoNormal | + | |
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | <tr style= | + | <tr style="height: 176.7pt;"> |
| − | <td | + | <td style="padding: 0cm 5.4pt; width: 415pt; height: 176.7pt;" valign="top" width="553"> |
| − | + | <p class="MsoNormal" style="text-indent: 0cm;"><b>B</b></p> | |
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-align: center; text-indent: 0cm; page-break-after: avoid;" align="center"><img style="width: 607px; height: 481px;" alt="" id="Diagramm 1156" src="https://static.igem.org/mediawiki/2010/0/0a/Freiburg10_MTT_96h_3d.png"></p> |
| − | <p class=MsoNormal | + | <p class="MsoCaption"><a name="_Ref275977314"><span lang="EN-US">Figure </span></a><span lang="EN-US">9</span><span lang="EN-US">: Effect of ganciclovir on HT1080 cell survival |
| − | + | 96 hours post infection. (A) two dimensional plot of cell survival (B) | |
| − | + | three-dimensional plot of ganciclovir, virus particles and cell survival.</span></p> | |
| − | <p class=MsoCaption><a name="_Ref275977314"><span lang=EN-US>Figure </span></a><span lang=EN-US>9</span><span lang=EN-US>: Effect of ganciclovir on HT1080 cell | + | |
| − | + | ||
| − | + | ||
| − | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
| − | </table> | + | </tbody></table> |
</div> | </div> | ||
| − | <p class=MsoNormal style= | + | <p class="MsoNormal" style="text-indent: 0cm;"><span style="font-size: 8pt; line-height: 200%;" lang="EN-US"> </span></p> |
| − | + | ||
| − | <p class=MsoNormal><span lang=EN-US>Data of MTT assay quantification are shown | + | <p class="MsoNormal"><span lang="EN-US">Data of MTT assay quantification are shown |
| − | in </span><span lang=EN-US>Figure 8</span><span lang=EN-US> and </span><span lang=EN-US>Figure 9</span><span lang=EN-US>. HT1080 cells were infected with | + | in </span><span lang="EN-US">Figure 8</span><span lang="EN-US"> and </span><span lang="EN-US">Figure 9</span><span lang="EN-US">. HT1080 cells were infected with |
| − | viral particles containing the mGMK_TK30 transgene. | + | viral particles containing the mGMK_TK30 transgene. 72 h- and 96 h post |
infection and addition of ganciclovir, cells were incubated with MTT. Changes | infection and addition of ganciclovir, cells were incubated with MTT. Changes | ||
in absorbance were measured and survival of cells plotted against ganciclovir | in absorbance were measured and survival of cells plotted against ganciclovir | ||
| − | concentration. </span><span lang=EN-US>Figure 8</span><span lang=EN-US>A | + | concentration. </span><span lang="EN-US">Figure 8</span><span lang="EN-US">A |
demonstrates the correlation between increasing ganciclovir concentrations and | demonstrates the correlation between increasing ganciclovir concentrations and | ||
percentage of cell survival. Furthermore, different virus particle | percentage of cell survival. Furthermore, different virus particle | ||
| − | concentrations were used for transduction. </span><span | + | concentrations were used for transduction. </span><span lang="EN-US">Figure 8</span><span lang="EN-US">B shows that the highest amount of |
| − | lang=EN-US>Figure 8</span><span lang=EN-US>B shows that the highest amount of | + | |
viral particles combined with the highest ganciclovir concentration led to | viral particles combined with the highest ganciclovir concentration led to | ||
significant HT1080 apoptosis 72 hours post transduction.</span></p> | significant HT1080 apoptosis 72 hours post transduction.</span></p> | ||
| − | <p class=MsoNormal><span lang=EN-US>Additionally 96 hours post infection cells | + | <p class="MsoNormal"><span lang="EN-US">Additionally, 96 hours post infection cells |
| − | were incubated with MTT and absorbance was quantified via spectrometry (</span><span lang=EN-US>Figure 9</span><span lang=EN-US>). Again, survival of HT1080 cells | + | were incubated with MTT and absorbance was quantified via spectrometry (</span><span lang="EN-US">Figure 9</span><span lang="EN-US">). Again, survival of HT1080 cells |
was plotted against increasing ganciclovir concentrations. </span></p> | was plotted against increasing ganciclovir concentrations. </span></p> | ||
| + | <h3><a name="_Toc275981847"><span lang="EN-US">Killing Non-transduced Tumor Cells | ||
| + | via Bystander Effect</span></a><span lang="EN-US"> </span></h3> | ||
| − | <p class=MsoNormal><span lang=EN-US> | + | <p class="MsoNormal"><span lang="EN-US">The bystander effect was first reported by </span><span lang="EN-US">Moolten (1986)</span><span lang="EN-US"> showing that prodrug |
| + | convertase negative cells surrounded by suicide enzyme positive cells did not | ||
| + | survive prodrug treatment. Besides efficient killing of targeted tumor cells, | ||
| + | neighboring, non-transduced cells are killed as well, providing an important | ||
| + | effect in cancer treatment. Since 5-Fluorouracil is soluble and can diffuse | ||
| + | into adjacent cells </span><span lang="EN-US">(Huber et al. 1993)</span><span lang="EN-US"> </span><span lang="EN-US">(Huber et al. 1994)</span><span lang="EN-US">, the bystander effect | ||
| + | was demonstrated using cytosine deaminase as gene of interest.</span></p> | ||
| − | < | + | <p class="MsoNormal"><span lang="EN-US"> </span></p> |
| − | <p class=MsoNormal><span lang=EN-US>Efficient and tissue-specific tumor killing | + | <div align="center"> |
| − | is one major challenge in cancer therapy </span><span | + | |
| − | lang=EN-US>(Black et al. 1996)</span><span lang=EN-US>. Gene-directed enzyme | + | <table class="MsoTableGrid" style="border: medium none ; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0"> |
| + | <tbody><tr> | ||
| + | <td style="padding: 0cm 5.4pt; width: 460.6pt;" valign="top" width="614"> | ||
| + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 614px; height: 262px;" alt="" id="Grafik 82" src="https://static.igem.org/mediawiki/2010/c/c0/Freiburg10_Bystander_effect.png"></p> | ||
| + | <p class="MsoCaption"><span lang="EN-US">Figure </span><span lang="EN-US">10</span><span lang="EN-US">: Schematic overview of the Bystander | ||
| + | effect.</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody></table> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US"> </span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">The aim was to investigate if the modified | ||
| + | AAV-2 is able to kill tumor cells with the cytosine deaminase (CD) as gene of interest. | ||
| + | The viral particles were produced according to the standard protocol using the | ||
| + | following plasmids:</span></p> | ||
| + | |||
| + | <p class="MsoListParagraphCxSpFirst" style="margin-left: 53.85pt; text-indent: -18pt;"><span style="font-family: Symbol;">·<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> | ||
| + | </span></span>pHelper</p> | ||
| + | |||
| + | <p class="MsoListParagraphCxSpMiddle" style="margin-left: 53.85pt; text-indent: -18pt;"><span style="font-family: Symbol;" lang="EN-US">·<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> | ||
| + | </span></span><span lang="EN-US">Rep/Cap: | ||
| + | pSB1C3_001_[AAV2]-Rep-VP123(ViralBrick-587KO-empty)_p5-TATAless</span></p> | ||
| + | |||
| + | <p class="MsoListParagraphCxSpLast" style="margin-left: 53.85pt; text-indent: -18pt;"><span style="font-family: Symbol;" lang="EN-US">·<span style="font-family: "Times New Roman"; font-style: normal; font-variant: normal; font-weight: normal; font-size: 7pt; line-height: normal; font-size-adjust: none; font-stretch: normal;"> | ||
| + | </span></span><span lang="EN-US">Gene of interest: | ||
| + | pSB1C3_[AAV2]-left-ITR_pCMV_betaglobin_CD_hGH_[AAV2]-right-ITR</span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">90 % confluent HT1080 cells were transduced | ||
| + | in T75 flasks with 9 ml of viral stock. In parallel, another T75 flask was | ||
| + | transduced with a viral stock packaged with the mVenus coding sequence to | ||
| + | assess transgene expression.</span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">24 hours before harvesting the | ||
| + | CD-transduced cells, two six well plates with 200.000 cells per well were | ||
| + | seeded. After 30 hours, mVenus expression was observed and the CD-transduced | ||
| + | HT1080 were harvested. To minimize the influence of viral particles in the | ||
| + | medium, the cells were washed four times with PBS. The cells were counted via a | ||
| + | Neubauer cell chamber and 100.000 cells per well of a six well plate were | ||
| + | seeded. Additionally, 100.000 of the CD-transduced cells were seeded onto previously | ||
| + | seeded untransduced HT1080.</span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">The incubation with the prodrug | ||
| + | 5-fluorocytosine (5-FC) was performed at a final concentration of 53 mM. | ||
| + | According to Fuchita <i>et al.,</i> this amount should be sufficient to show | ||
| + | the functionality of the CD </span><span lang="EN-US">(Fuchita et al. 2009b)</span><span lang="EN-US">. As demonstrated in </span><span lang="EN-US">Figure 11</span><span lang="EN-US">, cytotoxicity of 5-FC is | ||
| + | remarkable.</span></p> | ||
| + | |||
| + | <div align="center"> | ||
| + | |||
| + | <table class="MsoTableGrid" style="border: medium none ; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0"> | ||
| + | <tbody><tr style="height: 351pt;"> | ||
| + | <td style="padding: 0cm 5.4pt; width: 457.65pt; height: 351pt;" valign="top" width="610"> | ||
| + | <p class="MsoNormal" style="text-indent: 0cm; page-break-after: avoid;"><img style="width: 610px; height: 424px;" alt="" src="https://static.igem.org/mediawiki/2010/4/4c/Freiburg10_CD_ToxicEffect.png" id="Diagramm 5"></p> | ||
| + | <p class="MsoCaption" style="text-indent: 0cm;"><span lang="EN-US">Figure </span><span lang="EN-US">11</span><span lang="EN-US">: </span><span style="font-weight: normal;" lang="EN-US">Transduction of HT1080 with cytosine deaminase-packed viral | ||
| + | particles. 5-FC: 5-fluorocytosine (53 mM)</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody></table> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <p class="MsoNormal" style="text-indent: 0cm;"><span lang="EN-US"> </span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">After three days of incubation in 5-fluorocytosine, | ||
| + | the cells were washed, detached with Trypsin, centrifuged at 200 g for 5 min | ||
| + | followed by two washing steps with PBS and finally resuspended with 200 µl | ||
| + | DMEM. Living cells were then counted via Trypan blue staining.</span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">After this successful qualitative | ||
| + | demonstration of an AAV2-mediated cytosine deaminase treatment, the bystander | ||
| + | effect was quantified as well. The activated 5-FC molecules are able to diffuse | ||
| + | through the plasma membrane and thus effect cells that are not transduced (</span><span lang="EN-US">Figure 12</span><span lang="EN-US">). The bystander effect was tested with | ||
| + | untransduced HT1080 cells, which were mixed with the CD-transduced HT1080 cells</span><span lang="EN-US">.</span></p> | ||
| + | |||
| + | <div align="center"> | ||
| + | |||
| + | <table class="MsoTableGrid" style="border: medium none ; border-collapse: collapse;" border="0" cellpadding="0" cellspacing="0"> | ||
| + | <tbody><tr> | ||
| + | <td style="padding: 0cm 5.4pt; width: 460.6pt;" valign="top" width="614"> | ||
| + | <p class="MsoNormal" style="text-align: left; text-indent: 0cm; page-break-after: avoid;" align="left"><img style="width: 614px; height: 483px;" alt="" id="Diagramm 1158" src="https://static.igem.org/mediawiki/2010/8/8c/Freiburg10_CD_Bystander.png"></p> | ||
| + | <p class="MsoCaption" style="text-align: left;" align="left"><a name="_Ref275979941"><span lang="EN-US">Figure </span></a><span lang="EN-US">12</span><span lang="EN-US">: </span><span style="font-weight: normal;" lang="EN-US">Quantifying the bystander effect on | ||
| + | HT1080 cells 5-FC: 5-fluorocytosine (53 mM)</span></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody></table> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US"> </span></p> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">The cytosine deaminase expressing cells | ||
| + | have an obvious effect on the viability of the non-transduced cells. As shown | ||
| + | in the graph we were able to demonstrate that cytosine deaminase mediated cancer | ||
| + | cell death is also fatal for non-transduced cells in close proximity.</span></p> | ||
| + | |||
| + | <h3><a name="_Toc275981848"><span lang="EN-US">Conclusions</span></a></h3> | ||
| + | |||
| + | <p class="MsoNormal"><span lang="EN-US">Efficient and tissue-specific tumor killing | ||
| + | is one major challenge in cancer therapy </span><span lang="EN-US">(Black et al. 1996)</span><span lang="EN-US">. Gene-directed enzyme | ||
prodrug therapy (GDEPT) is based on the conversion of non-toxic substances into | prodrug therapy (GDEPT) is based on the conversion of non-toxic substances into | ||
toxic drugs resulting in tumor cell death. The iGEM team Freiburg_Bioware 2010 | toxic drugs resulting in tumor cell death. The iGEM team Freiburg_Bioware 2010 | ||
| − | provides several functional suicide genes within the Virus Construction Kit | + | provides several functional suicide genes within the Virus Construction Kit, thus |
offering a feasible and modular tool to the growing field of personalized | offering a feasible and modular tool to the growing field of personalized | ||
medicine and the iGEM community. We successfully demonstrated cancer cell death | medicine and the iGEM community. We successfully demonstrated cancer cell death | ||
| − | caused the modified fusion | + | caused by the introduction of cytosine deaminase and modified fusion genes |
consisting of guanylate and thymidine kinases.</span></p> | consisting of guanylate and thymidine kinases.</span></p> | ||
| − | <p class=MsoNormal><span lang=EN-US> | + | <p class="MsoNormal"><span lang="EN-US"> To prevent systemic toxic side effects of |
| − | conventional chemotherapy the iGEM team Freiburg_Bioware 2010 took a leap and | + | conventional chemotherapy, the iGEM team Freiburg_Bioware 2010 took a leap and |
efficiently retargeted the viral vector for directed suicide gene delivery | efficiently retargeted the viral vector for directed suicide gene delivery | ||
towards tumor cells. Capsid engineering was successfully demonstrated by the | towards tumor cells. Capsid engineering was successfully demonstrated by the | ||
| − | iGEM team Freiburg_Bioware 2010. Further details can be found | + | iGEM team Freiburg_Bioware 2010. Further details can be found under Results – |
| − | + | Targeting.</span></p> | |
| − | Targeting. | + | |
| − | <h3><a name="_Toc275981849"><span lang=EN-US>References</span></a></h3> | + | <h3><a name="_Toc275981849"><span lang="EN-US">References</span></a></h3> |
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Ardiani, A., Sanchez-Bonilla, M. & |
| − | font-family: | + | |
Black, M.E., 2010. Fusion enzymes containing HSV-1 thymidine kinase mutants and | Black, M.E., 2010. Fusion enzymes containing HSV-1 thymidine kinase mutants and | ||
guanylate kinase enhance prodrug sensitivity in vitro and in vivo. <i>Cancer | guanylate kinase enhance prodrug sensitivity in vitro and in vivo. <i>Cancer | ||
| Line 516: | Line 516: | ||
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2808426&tool=pmcentrez&rendertype=abstract.</span></p> | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2808426&tool=pmcentrez&rendertype=abstract.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Black, M.E. et al., 1996. Creation of |
| − | font-family: | + | |
drug-specific herpes simplex virus type 1 thymidine kinase mutants for gene | drug-specific herpes simplex virus type 1 thymidine kinase mutants for gene | ||
therapy. <i>Proceedings of the National Academy of Sciences of the United | therapy. <i>Proceedings of the National Academy of Sciences of the United | ||
| Line 523: | Line 522: | ||
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=39643&tool=pmcentrez&rendertype=abstract.</span></p> | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=39643&tool=pmcentrez&rendertype=abstract.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Fuchita, M. et al., 2009. Bacterial |
| − | font-family: | + | |
cytosine deaminase mutants created by molecular engineering show improved | cytosine deaminase mutants created by molecular engineering show improved | ||
5-fluorocytosine-mediated cell killing in vitro and in vivo. <i>Cancer research</i>, | 5-fluorocytosine-mediated cell killing in vitro and in vivo. <i>Cancer research</i>, | ||
| Line 530: | Line 528: | ||
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765227&tool=pmcentrez&rendertype=abstract.</span></p> | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765227&tool=pmcentrez&rendertype=abstract.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Fuchita, M. et al., 2009. Bacterial |
| − | font-family: | + | |
cytosine deaminase mutants created by molecular engineering show improved | cytosine deaminase mutants created by molecular engineering show improved | ||
5-fluorocytosine-mediated cell killing in vitro and in vivo. <i>Cancer research</i>, | 5-fluorocytosine-mediated cell killing in vitro and in vivo. <i>Cancer research</i>, | ||
| − | 69(11), 4791-9. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765227&tool=pmcentrez&rendertype=abstract.</span></p> | + | 69(11), 4791-9. Available at: |
| + | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765227&tool=pmcentrez&rendertype=abstract.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";">Greco, |
| − | O. & Dachs, G.U., 2001. </span><span | + | O. & Dachs, G.U., 2001. </span><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Gene directed enzyme/prodrug therapy of |
| − | font-family: | + | |
cancer: historical appraisal and future prospectives. <i>Journal of cellular | cancer: historical appraisal and future prospectives. <i>Journal of cellular | ||
physiology</i>, 187(1), 22-36. Available at: | physiology</i>, 187(1), 22-36. Available at: | ||
http://www.ncbi.nlm.nih.gov/pubmed/11241346.</span></p> | http://www.ncbi.nlm.nih.gov/pubmed/11241346.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";">Huber, |
| − | B.E. et al., 1993. </span><span | + | B.E. et al., 1993. </span><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">In vivo antitumor activity of 5-fluorocytosine on human |
| − | + | ||
colorectal carcinoma cells genetically modified to express cytosine deaminase. <i>Cancer | colorectal carcinoma cells genetically modified to express cytosine deaminase. <i>Cancer | ||
research</i>, 53(19), 4619-26. Available at: | research</i>, 53(19), 4619-26. Available at: | ||
http://www.ncbi.nlm.nih.gov/pubmed/8402637.</span></p> | http://www.ncbi.nlm.nih.gov/pubmed/8402637.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";">Huber, |
| − | B.E. et al., 1994. </span><span | + | B.E. et al., 1994. </span><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Metabolism of 5-fluorocytosine to 5-fluorouracil in |
| − | + | ||
human colorectal tumor cells transduced with the cytosine deaminase gene: | human colorectal tumor cells transduced with the cytosine deaminase gene: | ||
significant antitumor effects when only a small percentage of tumor cells | significant antitumor effects when only a small percentage of tumor cells | ||
| Line 559: | Line 554: | ||
http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=44594&tool=pmcentrez&rendertype=abstract.</span></p> | http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=44594&tool=pmcentrez&rendertype=abstract.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Moolten, F.L., 1986. Tumor chemosensitivity |
| − | font-family: | + | |
conferred by inserted herpes thymidine kinase genes: paradigm for a prospective | conferred by inserted herpes thymidine kinase genes: paradigm for a prospective | ||
cancer control strategy. <i>Cancer research</i>, 46(10), 5276-81. Available at: | cancer control strategy. <i>Cancer research</i>, 46(10), 5276-81. Available at: | ||
http://www.ncbi.nlm.nih.gov/pubmed/3019523.</span></p> | http://www.ncbi.nlm.nih.gov/pubmed/3019523.</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Roche, <i>Apoptosis , Cell Death and Cell |
| − | font-family: | + | |
Proliferation</i>,</span></p> | Proliferation</i>,</span></p> | ||
| − | <p style= | + | <p style="text-indent: 36pt;"><span style="font-size: 10pt; font-family: "Calibri","sans-serif";" lang="EN-US">Willmon, C.L., Krabbenhoft, E. & Black, |
| − | font-family: | + | |
M.E., 2006. A guanylate kinase/HSV-1 thymidine kinase fusion protein enhances | M.E., 2006. A guanylate kinase/HSV-1 thymidine kinase fusion protein enhances | ||
prodrug-mediated cell killing. <i>Gene therapy</i>, 13(17), 1309-12. Available | prodrug-mediated cell killing. <i>Gene therapy</i>, 13(17), 1309-12. Available | ||
at: http://www.ncbi.nlm.nih.gov/pubmed/16810197.</span></p> | at: http://www.ncbi.nlm.nih.gov/pubmed/16810197.</span></p> | ||
| − | <p | + | <p><span lang="EN-US"> </span></p> |
| − | + | ||
| − | + | ||
</html> | </html> | ||
Revision as of 00:11, 28 October 2010
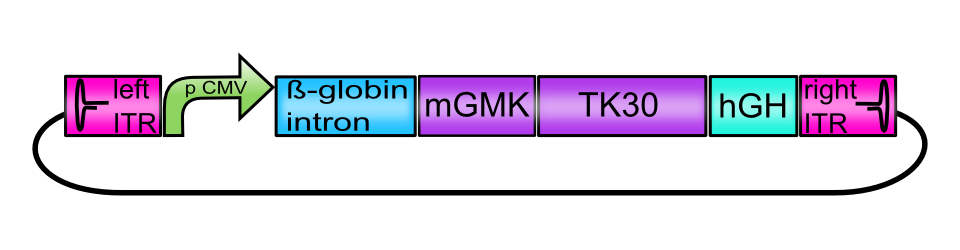
[AAV2]-left-ITR_pCMV_betaglobin_mGMK_TK30_hGH_[AAV2]-right-ITR
This composite BioBrick serves as an example for the possibilities of the Virus Construction Kit created by the iGEM Team Freiburg Bioware 2010. The has been assembled from 7 single BioBricks: A promoter, a transcription-enhancing sequence, a Gene of Interest that is itself a fusion protein, a polyadenylation sequence required for proper translation and inverted terminal repeats that are required for replication at both ends of the DNA molecule. Cotransfected into eukaryotic helper cell lines together with a plasmid encoding AAV Rep and Cap proteins and a pHelper plasmid encoding essential adenoviral genes, this BioBrick is being packaged into AAV capsids in form of a single-stranded DNA molecule and hence substitutes the AAV genome. AAV particles containing this Biobrick as a vector plasmid are replication-deficient and are therefore considered as biosafety level 1.
Contents
Successful Assembly of Vector Plasmids Carrying Suicide Genes via Cloning
Monitoring Efficient Tumor Killing by Phase-Contrast Microscopy
Quantitative Analysis of Cell Death by Flow Cytometry
Titrating Ganciclovir Concentrations for Efficient Cell Killing by Cytotoxicity Assays
Killing Untransduced Tumor Cells via Bystander Effect
Arming: Suicide Genes as GOIs
Introduction
Gene delivery using viral vectors to specifically target tumor cells gained increasing attention in the last years being efficient in combination with suicide gene approaches (Willmon et al. 2006). Several prodrug/enzyme combinations have been reported. Two systems - ganciclovir (GCV)/herpes simplex virus thymidine kinase (HSV-TK) (Ardiani et al. 2010) and 5-fluorocytosine/cytosine deaminase (CD) (Fuchita et al. 2009a) – have been widely used and their therapeutic benefit was demonstrated in preclinical studies (Greco & Dachs 2001). Adeno-associated viruses (AAV) as delivery vectors are commonly used in suicide gene therapy. The suicide gene flanked by the inverted terminal repeats (ITRs) is encapsulated into the virus particles and delivered to the target cells where suicide gene expression is mediated by cellular proteins.
The iGEM team Freiburg_Bioware 2010 provides both the cytosine deaminase (CD, BBa_K404112) and an improved guanylate kinase - thymidine kinase fusion gene (mGMK_TK, BBa_K404113) within the Virus Construction Kit as effective suicide genes. We demonstrate efficient and specific killing of tumor cells by enzymatic cytotoxicity assays, flow cytometry, as well as phase contrast microscopy. HT1080 cancer cell lines were transduced with directed viral particles containing the suicide genes packaged into the viral capsids.
Successful Assembly of Vector Plasmids Carrying Suicide Genes via Cloning
To create the functional vector plasmids, assembly of the constructs carrying the suicide genes was performed following the BioBrick Standard Assembly. All plasmids contain the enhancer-element human beta-globin intron (BBa_K404107) and the human growth hormone terminator signal (hGH, BBa_K404108) flanked by the inverted terminal repeats (ITRs, BBa_K404100 and BBa_K404101). Assembled suicide genes are either under the control of the CMV promoter (BBa_J52034) or the tumor-specific telomerase promoter phTERT (BBa_K404106).
|
Figure 1: BioBrick-compatible assembly of functional vector plasmids containing the suicide genes. The schematic figure shows the cloning strategy of the guanylate kinase – thymidine kinase fusion gene (mGMK_TK30). |
To enable modularization of the thymidine kinase mutants TK30 and SR39 (BBa_K404109 and BBa_K404110) according to the BioBrick standard, the fusion genes mGMK_TK30 and mGMK_SR39 (BBa_K404113 and BBa_K404315) and CD (BBa_K404112) were modified using the QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene) for deletion of iGEM RFC10 pre- and suffix restriction sites. Figure 1 demonstrates one example of successful deletion of a PstI restriction site located within the mGMK_TK30 sequence at position 3109. A point mutation was introduced replacing the nucleotide G by A, resulting in the deletion of the restriction site while maintaining the encoded amino acid glutamine. Exchange of guanine to adenine was confirmed by sequencing (Figure 2).
|
Figure 2: Cytosine to guanine exchange by site-directed mutagenesis using QuikChange Lightning Kit provided by Stratagene was successful as demonstrated by (A) test digestion linearizing the plasmid with PstI and (B) by sequencing. |
Furthermore, assembly of BioBrick-compatible vector plasmids was performed. An example for the last assembly step of mGMK_TK30 and hGH_rITR is shown in Figure 3. The plasmids were digested with both XbaI and PstI (New England Biolabs, Insert: BBa_K404116: hGH_rITR) or SpeI and PstI (Vector) and loaded on an agarose gel. As demonstrated in the preparative gel in Figure 3, the expected bands were detected under UV light and the extracted, subsequently purified DNA was successfully ligated and transformed into E. coli. Each assembly step for producing the BioBricks was conducted following the iGEM BioBrick standard.
|
Figure 3: Assembly of mGMK_TK30 (vector molecule, in pSB1C3) and hGH-terminator_rightITR (insert molecule). The digested fragments were visualized under UV-light and correspond to the expected sizes. |
Monitoring Efficient Tumor Killing by Phase-Contrast Microscopy
Tumor cells, transduced with viral particles encapsidating the effector constructs containing the mGMK_TK30 driven by the CMV promoter, were cultured both in presence and absence of ganciclovir (Roche). Morphological changes were monitored via phase-contrast microscopy for 48 hours post infection. As it can be seen in Figure 4, non-transduced tumor cells treated with ganciclovir and transduced cells without ganciclovir did not show significant cell ablation. In contrast, transduced cells expressing the guanylate kinase - thymidine kinase fusion protein showed significant cell death after incubation with ganciclovir for 48 hours post infection.
|
A |
B |
|
C
|
D |
|
Figure 4: Qualitative analysis of cell death induced by conversion of ganciclovir to ganciclovir-triphosphate by virus-delivered guanylate - thymidine kinase (mGMK_TK30). A: Non-transduced HT1080 cells incubated in the presence of ganciclovir not exhibiting cell death. B: Untreated transduced HT1080 cells showing a high survival rate. C: HT1080 cells transduced with 300 µL viral particles and incubated with ganciclovir leading to tumor cell ablation. D: HT1080 cells transduced with 600 µL viral particles and incubated with ganciclovir leading to ablation of tumor cells. |
|
Suicide gene therapy is based on the localized conversion of non-toxic prodrugs to toxic substances (Greco & Dachs 2001), promoting cell death in the tumor tissue (Figure 5). Directed gene delivery is achieved by using recombinant viral vectors as provided by the iGEM team Freiburg_Bioware 2010 within the Virus Construction Kit.
|
Figure 5: Overview of the suicide gene therapy approach. Non-toxic prodrugs are converted into toxic effector molecules leading to cell death of the tumor cells. |
Non-transduced cells can survive in the presence of ganciclovir since the prodrug is not toxic for these cells (Figure 4A). The demonstration that transduced cells are viable in the absence of ganciclovir confirms that cell killing is indeed induced by combination of the delivered thymidine kinase and treatment with ganciclovir. Viral particles encapsidating the suicide construct mGMK_TK30 are efficient in directed gene delivery, thus leading to cell death of transduced cells due to overexpression of mGMK_TK30 and prodrug conversion. The cell-toxic ganciclovir-triphosphate is incorporated into the nascent DNA chain leading to replication termination and finally resulting in death of dividing cells.
Quantitative Analysis of Cell Death by Flow Cytometry
Quantitative analysis of the cytotoxic effect induced by mGMK_TK30 was first conducted by flow cytometry analysis 72 hours post transduction. HT1080 cells were stained with 7-AAD and Annexin V. 7-AAD intercalates in double-stranded DNA after penetrating cell membranes of dead cells, whereas Annexin V specifically binds phosphatidylserine which is only accessible during apoptosis. Figure 6 demonstrates the relation between cell death and ganciclovir concentration.
|
|
|
|
Figure 6: Flow cytometry data analysis. A: Gating non-transduced HT1080 cells (control). B: Non-transduced cells without staining plotted against 7-AAD. C: Gating non-transduced cells stained with 7-AAD. D: Non-transduced, 7-AAD-stained cells plotted against 7-AAD. E: Gating transduced cells (GOI: mGMK_TK30) treated with 485 µM ganciclovir. F: Gated, Annexin-V stained cells plotted against AnnV-2 Log. G: Gated cells plotted against 7-AAD H: Gated, 7-AAD and Annexin-V stained cells plotted against 7-AAD and Annexin-V. Gate R19 comprised Annexin-V and 7-AAD positive cells. |
|
|
Figure 7: Quantification of flow cytometry data provided in Figure 6. With increasing ganciclovir concentration, the survival rate of cells decreases. 60 % of HT1080 cells treated with 4.85 mM ganciclovir show tumor ablation, however even lower amounts of ganciclovir lead to significant cell death. |
The effect of different ganciclovir concentrations on transduced HT1080 sarcoma cells was investigated. Transduction was performed with recombinant viral particles encapsidating the mGMK_TK30 prodrug gene. 72 hours post infection, cells were stained with 7-AAD and Annexin V. As Figure 7 shows, the fraction of killed transduced cells is proportional to the applied ganciclovir concentration.
Titrating Ganciclovir Concentrations for Efficient Cell Killing by Cytotoxicity Assays
Further analysis of the cytotoxic effect induced by thymidine kinase converting ganciclovir to the toxic anti-metabolite has been performed using MTT assays. 3-(4,5-Dimethylthiazol-2-yl)-2,5diphenyltetrazolium bromide), also known as MTT, is a yellow-colored tetrazole, which is reduced to purple insoluble formazan in the presence of NADH and NADPH (Roche n.d.). Colorimetric analysis can be carried out via spectrometry. Different tumor cell lines, HT1080 and A431, were transduced with the recombinant viruses carrying the linear DNA construct coding for mGMK-TK30 regulated by the CMV promoter and subsequently treated with ganciclovir. 48 and 72 hours post infection, cells were incubated with MTT and fresh DMEM. After cell lysis by DMSO, absorbance of formazan at 570 nm was quantified using a Tecan Sunrise plate reader.
|
A |
|
B
Figure 8: Effect of ganciclovir on HT1080 cell survival 72 hours post infection as (A) two dimensional plot of survival of cells and (B) three-dimensional plot of ganciclovir, virus particles and cell survival. |
|
A
|
|
B
Figure 9: Effect of ganciclovir on HT1080 cell survival 96 hours post infection. (A) two dimensional plot of cell survival (B) three-dimensional plot of ganciclovir, virus particles and cell survival. |
Data of MTT assay quantification are shown in Figure 8 and Figure 9. HT1080 cells were infected with viral particles containing the mGMK_TK30 transgene. 72 h- and 96 h post infection and addition of ganciclovir, cells were incubated with MTT. Changes in absorbance were measured and survival of cells plotted against ganciclovir concentration. Figure 8A demonstrates the correlation between increasing ganciclovir concentrations and percentage of cell survival. Furthermore, different virus particle concentrations were used for transduction. Figure 8B shows that the highest amount of viral particles combined with the highest ganciclovir concentration led to significant HT1080 apoptosis 72 hours post transduction.
Additionally, 96 hours post infection cells were incubated with MTT and absorbance was quantified via spectrometry (Figure 9). Again, survival of HT1080 cells was plotted against increasing ganciclovir concentrations.
Killing Non-transduced Tumor Cells via Bystander Effect
The bystander effect was first reported by Moolten (1986) showing that prodrug convertase negative cells surrounded by suicide enzyme positive cells did not survive prodrug treatment. Besides efficient killing of targeted tumor cells, neighboring, non-transduced cells are killed as well, providing an important effect in cancer treatment. Since 5-Fluorouracil is soluble and can diffuse into adjacent cells (Huber et al. 1993) (Huber et al. 1994), the bystander effect was demonstrated using cytosine deaminase as gene of interest.
|
Figure 10: Schematic overview of the Bystander effect. |
The aim was to investigate if the modified AAV-2 is able to kill tumor cells with the cytosine deaminase (CD) as gene of interest. The viral particles were produced according to the standard protocol using the following plasmids:
· pHelper
· Rep/Cap: pSB1C3_001_[AAV2]-Rep-VP123(ViralBrick-587KO-empty)_p5-TATAless
· Gene of interest: pSB1C3_[AAV2]-left-ITR_pCMV_betaglobin_CD_hGH_[AAV2]-right-ITR
90 % confluent HT1080 cells were transduced in T75 flasks with 9 ml of viral stock. In parallel, another T75 flask was transduced with a viral stock packaged with the mVenus coding sequence to assess transgene expression.
24 hours before harvesting the CD-transduced cells, two six well plates with 200.000 cells per well were seeded. After 30 hours, mVenus expression was observed and the CD-transduced HT1080 were harvested. To minimize the influence of viral particles in the medium, the cells were washed four times with PBS. The cells were counted via a Neubauer cell chamber and 100.000 cells per well of a six well plate were seeded. Additionally, 100.000 of the CD-transduced cells were seeded onto previously seeded untransduced HT1080.
The incubation with the prodrug 5-fluorocytosine (5-FC) was performed at a final concentration of 53 mM. According to Fuchita et al., this amount should be sufficient to show the functionality of the CD (Fuchita et al. 2009b). As demonstrated in Figure 11, cytotoxicity of 5-FC is remarkable.
|
Figure 11: Transduction of HT1080 with cytosine deaminase-packed viral particles. 5-FC: 5-fluorocytosine (53 mM) |
After three days of incubation in 5-fluorocytosine, the cells were washed, detached with Trypsin, centrifuged at 200 g for 5 min followed by two washing steps with PBS and finally resuspended with 200 µl DMEM. Living cells were then counted via Trypan blue staining.
After this successful qualitative demonstration of an AAV2-mediated cytosine deaminase treatment, the bystander effect was quantified as well. The activated 5-FC molecules are able to diffuse through the plasma membrane and thus effect cells that are not transduced (Figure 12). The bystander effect was tested with untransduced HT1080 cells, which were mixed with the CD-transduced HT1080 cells.
|
Figure 12: Quantifying the bystander effect on HT1080 cells 5-FC: 5-fluorocytosine (53 mM) |
The cytosine deaminase expressing cells have an obvious effect on the viability of the non-transduced cells. As shown in the graph we were able to demonstrate that cytosine deaminase mediated cancer cell death is also fatal for non-transduced cells in close proximity.
Conclusions
Efficient and tissue-specific tumor killing is one major challenge in cancer therapy (Black et al. 1996). Gene-directed enzyme prodrug therapy (GDEPT) is based on the conversion of non-toxic substances into toxic drugs resulting in tumor cell death. The iGEM team Freiburg_Bioware 2010 provides several functional suicide genes within the Virus Construction Kit, thus offering a feasible and modular tool to the growing field of personalized medicine and the iGEM community. We successfully demonstrated cancer cell death caused by the introduction of cytosine deaminase and modified fusion genes consisting of guanylate and thymidine kinases.
To prevent systemic toxic side effects of conventional chemotherapy, the iGEM team Freiburg_Bioware 2010 took a leap and efficiently retargeted the viral vector for directed suicide gene delivery towards tumor cells. Capsid engineering was successfully demonstrated by the iGEM team Freiburg_Bioware 2010. Further details can be found under Results – Targeting.
References
Ardiani, A., Sanchez-Bonilla, M. & Black, M.E., 2010. Fusion enzymes containing HSV-1 thymidine kinase mutants and guanylate kinase enhance prodrug sensitivity in vitro and in vivo. Cancer gene therapy, 17(2), 86-96. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2808426&tool=pmcentrez&rendertype=abstract.
Black, M.E. et al., 1996. Creation of drug-specific herpes simplex virus type 1 thymidine kinase mutants for gene therapy. Proceedings of the National Academy of Sciences of the United States of America, 93(8), 3525-9. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=39643&tool=pmcentrez&rendertype=abstract.
Fuchita, M. et al., 2009. Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo. Cancer research, 69(11), 4791-9. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765227&tool=pmcentrez&rendertype=abstract.
Fuchita, M. et al., 2009. Bacterial cytosine deaminase mutants created by molecular engineering show improved 5-fluorocytosine-mediated cell killing in vitro and in vivo. Cancer research, 69(11), 4791-9. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765227&tool=pmcentrez&rendertype=abstract.
Greco, O. & Dachs, G.U., 2001. Gene directed enzyme/prodrug therapy of cancer: historical appraisal and future prospectives. Journal of cellular physiology, 187(1), 22-36. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11241346.
Huber, B.E. et al., 1993. In vivo antitumor activity of 5-fluorocytosine on human colorectal carcinoma cells genetically modified to express cytosine deaminase. Cancer research, 53(19), 4619-26. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8402637.
Huber, B.E. et al., 1994. Metabolism of 5-fluorocytosine to 5-fluorouracil in human colorectal tumor cells transduced with the cytosine deaminase gene: significant antitumor effects when only a small percentage of tumor cells express cytosine deaminase. Proceedings of the National Academy of Sciences of the United States of America, 91(17), 8302-6. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=44594&tool=pmcentrez&rendertype=abstract.
Moolten, F.L., 1986. Tumor chemosensitivity conferred by inserted herpes thymidine kinase genes: paradigm for a prospective cancer control strategy. Cancer research, 46(10), 5276-81. Available at: http://www.ncbi.nlm.nih.gov/pubmed/3019523.
Roche, Apoptosis , Cell Death and Cell Proliferation,
Willmon, C.L., Krabbenhoft, E. & Black, M.E., 2006. A guanylate kinase/HSV-1 thymidine kinase fusion protein enhances prodrug-mediated cell killing. Gene therapy, 13(17), 1309-12. Available at: http://www.ncbi.nlm.nih.gov/pubmed/16810197.
ITRs

The inverted terminal repeat structures can be subdivided into several palindromic motives: A and A’ form a stem loop which encases B and B’ as well as C and C’. Those motives form both arms of the T-shaped structure. The functional motives on the ITR are two regions that bind Rep 68/78, called Rep-binding elements (RBE on the stem and RBE’ on the B arm) and the terminal resolution site (trs) in which the rep proteins introduce single-stranded nicks. The 3’ OH end of the A motive acts as a primer for DNA replication (Im & Muzyczka, 1990) (Lusby, Fife, & Berns, 1980).
CMV promoter
Beta-globin-intron


The beta-globin intron BioBrick consists of a partial chimeric CMV promoter followed by the intron II of the beta-globin gene. The 3´end of the intron is fused to the first 25 bases of human beta globin gene exon 3. The beta globin intron BioBrick is assumed to enhance eukaryotic gene expression (Nott, Meislin, & Moore, 2003). As shown in Figure 9 and Figure 10 the vectorplasmid missing the beta-globin intron showed a negligible difference in mVenus expression compared to viral genomes containing the beta-globin intron. Considering these results and taking into account that a constant volume of viral particles has been used for transduction, the difference between the construct containing and lacking the beta-globin intron is minimal. Since packaging efficiency of the AAV-2 decreases with increasing sizes of the insert (Dong, Fan, & Frizzell, 1996), the iGEM team Freiburg_Bioware suggests using the beta-globin intron in dependence on the size of your transgene.
mGMK_TK30 fusion protein
The thymidine kinase mutant TK30 contains six modified amino acids (Black, Newcomb, Wilson, & Loeb, 1996) created in a first screening showing enhanced affinity for gancivlocir and acyclovir, but reduced specificity for its natural substrate thymidine.
As efficient tumor killing and therefore ganciclovir activation is essential for successful tumor ablation, further improvements were conducted. Overexpression of transgenic thymidine kinase leads to accumulation of non-toxic intermediates, which cannot be phosphorylated sufficiently by endogenous guanylate kinase, the second enzyme in the salvage pathway of nucleotides.
Overcoming this bottleneck was accomplished by fusing the mouse guanylate kinase (mGMK) to the N-terminus of TK30 mutant creating a fusion protein (mGMK_TK30) with enhanced GCV/ACV sensitivity in vitro and in vivo (Ardiani, Sanchez-Bonilla, & Black, 2010) and improved bystander activity. The effect of non-transfected tumor cell killing upon transfer of toxic metabolites through gap junctional intercellular communication (GJIC) or immune-mediated tumor ablation is essential in suicide gene therapy (Pope, 1997). GCV-triphosphate is mainly transported through the central pore formed between connexin proteins from neighboring cells (Gentry, Im, Boucher, Ruch, & Shewach, 2005), but immune-induced bystander effect seems to be likely as well (Grignet-Debrus, Cool, Baudson, Velu, & Calberg-Bacq, 2000).
hGH
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1319
Illegal NgoMIV site found at 1977
Illegal NgoMIV site found at 2882
Illegal AgeI site found at 3011 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 3413
Illegal SapI.rc site found at 1369