Difference between revisions of "Part:BBa K801091"
ProfessorLi (Talk | contribs) (→Usage and Biology) |
|||
| Line 50: | Line 50: | ||
<html> | <html> | ||
<div style="display:flex; flex-direction: column; align-items: center;"> | <div style="display:flex; flex-direction: column; align-items: center;"> | ||
| − | <img src="https://static.igem.wiki/teams/5013/wiki/part/emphasis-basic-components-1-pal-gene-old-part-contribution/ | + | <img src="https://static.igem.wiki/teams/5013/wiki/part/emphasis-basic-components-1-pal-gene-old-part-contribution/2023-10-12-11-48-21.png" style="width: 500px;margin: 0 auto" /> |
<p style="font-size: 98%; line-height: 1.4em;"> Figure1 The design of PAL. | <p style="font-size: 98%; line-height: 1.4em;"> Figure1 The design of PAL. | ||
</p > | </p > | ||
Revision as of 09:47, 12 October 2023
phenylalanine ammonia lyase (PAL) coding region
Coding region for the enzyme phenylalanine ammonia lyase (PAL) from Rhodosporidium toruloides
Background and priniciples
Usage and Biology
Inhibition of the metabolic activation of procarcinogens:
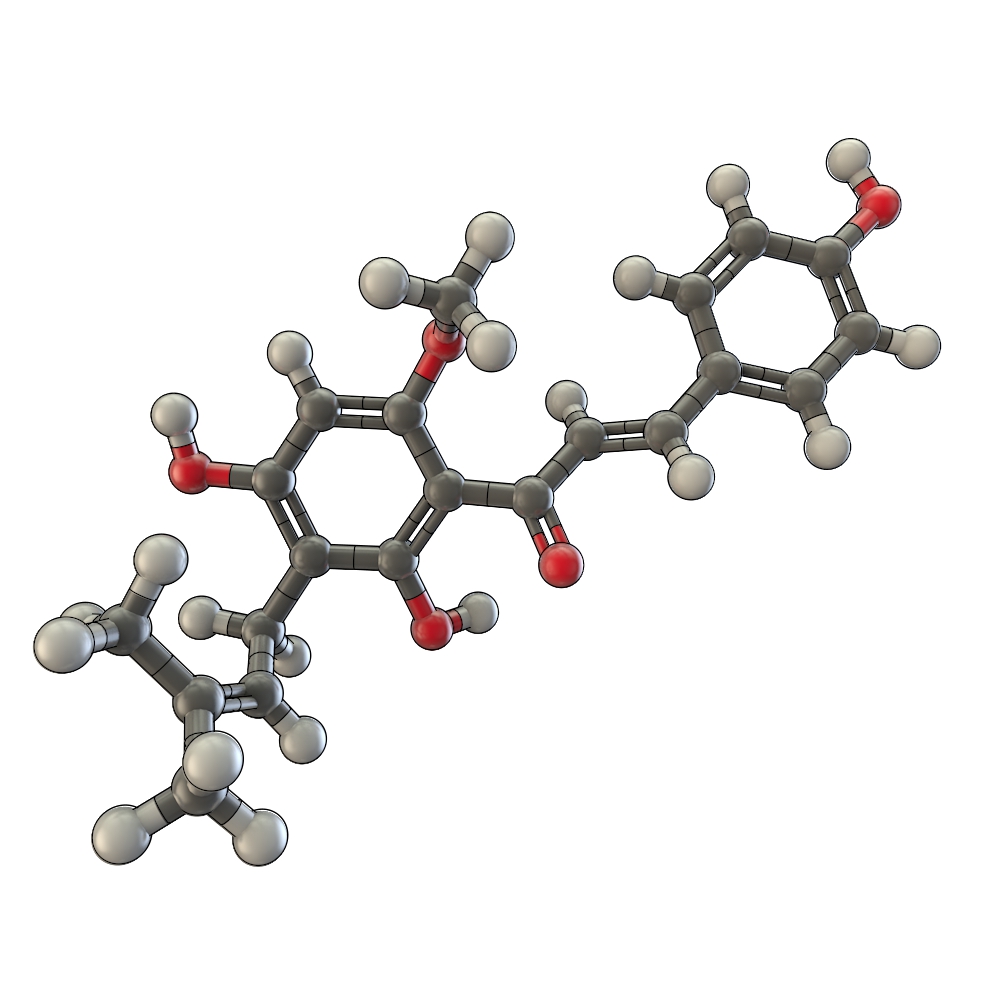
2-amino-3-methylimidazo[4,5-f]quinolone, found in cooked meat, verified as a procarcinogen in an ames salmonella mutagenicity test. The inhibition is probably a result of an inhibition of the cytochrome P 450 enzymes Cyp1A1, Cyp1B1 and Cyp1A2 (phase 1 enzymes). But in order to achieve a clear inhibition, plasma concentrations of 1 µM would be necessary. In a study with male rats oral administration of xanthohumol (50 mg/kg) led to concentration maximums of 65 -180 nM after 4 h. Improved resorption of xanthohumol could be a possible target for innovation [[http://www.ncbi.nlm.nih.gov/pubmed/11240137 Yilmazer et al. 2001a], [http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al. 2000b], [http://www.ncbi.nlm.nih.gov/pubmed/10752639 Henderson et al., 2000], [http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002]].
Induction of carcinogen-detoxifying enzymes (phase 2 enzymes):
P450-activated carcinogens get conjugated to endogenous ligands (gluthathione, glucoronic acid, acetate and sulfate) by phase 2 enzymes to facilitate excretion. Therefore the induction of phase 2 enzymes should enhance the protection against carcinogenesis. Xanthohumol cat concentrations of 2.1-10.1 µM could induce quinone reductase (detoxification of quinones by conversion to hydroquinones which can be conjugated) in hepatoma Hepa 1c1c7 cells. It was shown that xanthohumol could selectively induce quinone reductase without causing a transcriptional activation of Cyp1A1 [[http://www.ncbi.nlm.nih.gov/pubmed/11038156 Miranda et al., 2000c], [http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002]].
Inhibition of tumor growth at an early stage:
Xanthohumol showed an inhibition of the proliferation of breast cancer (MCF-7) and ovarian cancer (A-2780) in vitro at IC50 values of 13 and 0.52 µM http://www.ncbi.nlm.nih.gov/pubmed/10418944 Miranda ''et al.'', 1999. Furthermore xanthohumol can inhibit the endogenous prostaglandin synthesis through inhibition of cyclooxygenase (COX-1 and COX-2) with IC50 values of 17 and 42 µM. An increased prostaglandin production has been associated with the uncontrolled proliferation of tumor cells http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser ''et al.'', 2002. Pharmacokinetic studies for xanthohumol based on beverages with an xanthohumol content of 50 mg/l in humans are part of actual research activities.
Antioxidant activities:
Xanthohumol at 5 µM decreased conjugated diene formation as a measure for lipid peroxidation by more than 70 % after 5 h of incubation in an in vitro assay (protection of LDL from Cu2+ induced oxidation). Furthermore xanthohumol was shown to scavenge hydroxyl-, peroxyl- and superoxide anion radicals http://www.ncbi.nlm.nih.gov/pubmed/11038156 Miranda ''et al.'', 2000c. </div>
Biosynthesis
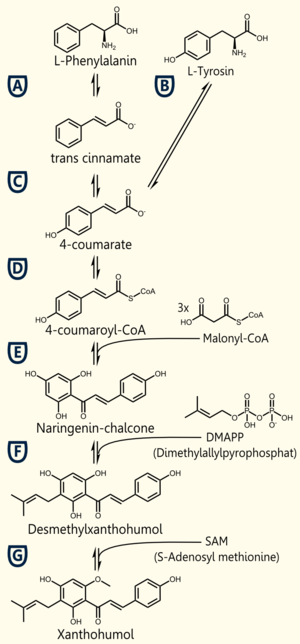
The biosynthetic pathway of 4-coumaroyl-coenzyme A starts with the conversion of L-Phenylalanine to cinnamate, being catalyzed by phenylalanin ammonia lyase (PAL) [A]. PAL also shows activity in converting tyrosine to p-coumarate, but with a lower efficiency [B]. The cinnamate 4-hydroxylase (C4H) catalyzes the synthesis of p-hydroxycinnamate from cinnamate and 4-coumarate [C]: CoA ligase (4CL) converts p-coumarate to its coenzyme-A ester, activating it for reaction with malonyl CoA [D] [Trantas et al., 2009].
The flavonoid biosynthetic pathway starts with the condensation of one molecule of 4-coumaroyl-CoA and three molecules of malonyl-CoA, yielding naringenin chalcone. This reaction is carried out by the enzyme chalcone synthase (CHS) [E]. Chalcone is isomerised to a flavanone by the enzyme chalcone flavanone isomerase (CHI). From these central intermediates, the pathway diverges into several side branches, each resulting in a different class of flavonoids, such as xanthohumol.
There are 5 enzymes necessary for the biosynthesis of xanthohumol ([http://biocyc.org/META/NEW-IMAGE?type=NIL&object=PWY-5135 MetaCyc]):
Enzyme [A]: PAL = phenylalanine ammonia lyase: L-phenylalanin --> trans-cinnamate
Enzyme [D]: 4CL = 4-coumarate - coenzym A ligase: 4-coumarate --> 4-coumaroyl-CoA
Enzyme [E]: CHS = naringenin - chalcone synthase: 4-coumaroyl-CoA --> naringeninchalcone
Enzyme [F]: APT = aromatic prenyltransferase: naringeninchalcone --> desmethylxanthohumol
Enzyme [G]: OMT1 = chalcone O-methyltransferase: desmethylxanthohumol --> xanthohumol
Jiang et al succeeded in the biosynthesis of several flavonoids in Saccharomyces cerevisiae by the assembly of a plasmid containing three required enzymes (pKS2µHyg-PAL-4CL-CHS) and thereby showed the proof of principle. The activity of each enzyme was demonstrated and the presence of naringenin, which forms the product of the three enzymes( PAL, 4CL, CHS), was shown.
http://www.ncbi.nlm.nih.gov/pubmed/14704995 Jiang and Morgan, 2004
Squirrel-CHN 2023
Description
Phenylalanine ammonia lyase (PAL) is an essential enzyme that catalyzes the deamination, the removal of an amino group, of phenylalanine (Phe) that PKU patients cannot process.
Usage and Biology
We express the pal gene by utilizing the pLac and the ribosome binding site B0034. The pET23b vector will be employed, and the engineered plasmid will be introduced into E.coli Rosetta for efficient expression.

Figure1 The design of PAL.

Figure2 Gel electrophoresis of the pal gene.
Characterization
By measuring the Phe content and TCA content after expressing the engineered strains, the degradation ability of PAL was evaluated. The strains were resuspended in 1 mL of Phe experimental buffer (M9 0.5% glucose with 1 mM Phe) to an OD600 of 0.1, and the Phe content was determined using a phenylalanine ELISA kit. Based on the Phe content measurement experiment, we used an microplate reader reader to measure OD290 and calculate the concentration of TCA. The results are shown in Figure 4. After expressing PAL, the Phe content decreased while the TCA content significantly increased, indicating that PAL can effectively degrade Phe into TCA.

Figure3 Schematic diagram of pal.

Figure 4 PAL degradation capability.
Additionally, we also validated the effect of environmental pH on the metabolic capacity of PAL. As shown in Figure 5, when the environmental pH value ranged from 5 to 8, the content of TCA increased with the increase in pH value. Therefore, this experiment can demonstrate that within the pH range of 5 to 8, the activity of PAL increases with the increase in pH value, with pH 8 being the optimal pH.

Figure 5 Testing the impact of pH environment on the degradation ability of PAL.
Potential application directions
Using synthetic biology techniques to produce enzymes that can degrade Phe, which can be applied in the treatment of phenylketonuria, provides new ideas and methods for this disease.
References
Bagal, U. R., Leebens-Mack, J. H., Lorenz, W. W., & Dean, J. F. (2012). The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. BMC genomics, 13 Suppl 3(Suppl 3), S1. https://doi.org/10.1186/1471-2164-13-S3-S1
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2090
Illegal BamHI site found at 352
Illegal XhoI site found at 403
Illegal XhoI site found at 466
Illegal XhoI site found at 484
Illegal XhoI site found at 562
Illegal XhoI site found at 763
Illegal XhoI site found at 1006
Illegal XhoI site found at 1753 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 922
Illegal NgoMIV site found at 1168
Illegal NgoMIV site found at 1336
Illegal NgoMIV site found at 1479
Illegal NgoMIV site found at 1813 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 463
Illegal BsaI site found at 793
Illegal BsaI site found at 799
Illegal BsaI.rc site found at 1924
Illegal SapI site found at 753
References
- http://www.ncbi.nlm.nih.gov/pubmed/12481418 Gerhauser et al., 2002 Gerhauser, C., Alt, A., Heiss, E., Gamal-Eldeen, A., Klimo, K., Knauft, J., Neu- mann, I., Scherf, H.-R., Frank, N., Bartsch, H., and Becker, H. (2002). Cancer chemopreventive activity of xanthohumol, a natural product derived from hop. Mol Cancer Ther, 1(11):959–69.
- http://www.ncbi.nlm.nih.gov/pubmed/10752639 Henderson et al., 2000 Henderson, M. C., Miranda, C. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000). In vitro inhibition of human p450 enzymes by prenylated flavonoids from hops, humulus lupulus. Xenobiotica, 30(3):235–51.
- http://www.ncbi.nlm.nih.gov/pubmed/14704995 Jiang and Morgan, 2004 Jiang, H. and Morgan, J. A. (2004). Optimization of an in vivo plant p450 monooxygenase system in Saccharomyces cerevisiae. Biotechnol Bioeng, 85(2):130–7.
- http://www.ncbi.nlm.nih.gov/pubmed/10737704 Miranda et al., 2000a Miranda, C. L., Aponso, G. L., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2000a). Prenylated chalcones and flavanones as inducers of quinone reductase in mouse hepa 1c1c7 cells. Cancer Lett, 149(1-2):21–9.
- http://www.ncbi.nlm.nih.gov/pubmed/10418944 Miranda et al., 1999 Miranda, C. L., Stevens, J. F., Helmrich, A., Henderson, M. C., Rodriguez, R. J., Yang, Y. H., Deinzer, M. L., Barnes, D. W., and Buhler, D. R. (1999). Antiproliferative and cytotoxic effects of prenylated flavonoids from hops (Humulus lupulus) in human cancer cell lines. Food Chem Toxicol, 37(4):271–85.
- http://www.ncbi.nlm.nih.gov/pubmed/10995285 Miranda et al., 2000b Miranda, C. L., Stevens, J. F., Ivanov, V., McCall, M., Frei, B., Deinzer, M. L., and Buhler, D. R. (2000b). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J Agric Food Chem, 48(9):3876–84.
- http://www.ncbi.nlm.nih.gov/pubmed/11038156 Miranda et al., 2000c Miranda, C. L., Yang, Y. H., Henderson, M. C., Stevens, J. F., Santana-Rios, G., Deinzer, M. L., and Buhler, D. R. (2000c). Prenylflavonoids from hops inhibit the metabolic activation of the carcinogenic heterocyclic amine 2-amino-3-methylimidazo[4, 5-f]quinoline, mediated by cdna-expressed human cyp1a2. Drug Metab Dispos, 28(11):1297–302.
- http://www.ncbi.nlm.nih.gov/pubmed/19631278 Trantas et al., 2009 Trantas, E., Panopoulos, N., and Ververidis, F. (2009). Metabolic engineering of the complete pathway leading to heterologous biosynthesis of various flavonoids and stilbenoids in Saccharomyces cerevisiae. Metab Eng, 11(6):355–66.
- http://www.ncbi.nlm.nih.gov/pubmed/11240137 Yilmazer et al., 2001a Yilmazer, M., Stevens, J. F., and Buhler, D. R. (2001a). In vitro glucuronidation of xanthohumol, a flavonoid in hop and beer, by rat and human liver microsomes. FEBS Lett, 491(3):252–6.
- http://www.ncbi.nlm.nih.gov/pubmed/11181488 Yilmazer et al., 2001b Yilmazer, M., Stevens, J. F., Deinzer, M. L., and Buhler, D. R. (2001b). In vitro biotransformation of xanthohumol, a flavonoid from hops (Humulus lupulus), by rat liver microsomes. Drug Metab Dispos, 29(3):223–31.