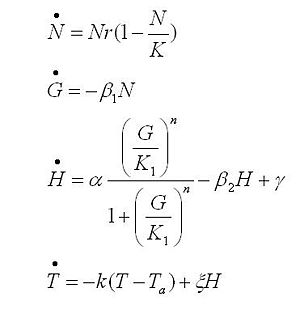Difference between revisions of "Part:BBa K141001"
| Line 64: | Line 64: | ||
<partinfo>BBa_K141001 parameters</partinfo> | <partinfo>BBa_K141001 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | ==Contribution: NUDT_CHINA 2023== | ||
| + | In our project for iGEM2023, we aims to design a system that can effectively deliver the uncoupling protein UCP1 into adipocytes via PVCs.Herein,one composite part from the registry was used to generate the plasmid for functionality of EGFP-UCP1 Payload Protein in HEK-293T Cells .Remarkably,this part,uncoupling protein UCP1, was submitted by team iGEM08_Valencia.However, we found important information missing on the registry pages of this part. Therefore, we made efforts to provide experimental validation data and our protocol to enable the PVC-based delivery of the UCP1 part into target cells. | ||
| + | <html> | ||
| + | |||
| + | <figure class="figure"> | ||
| + | <img src="https://static.igem.wiki/teams/4960/wiki/basic-part/functionality-of-ucp1-based-payload-protein-in-hek-293t-cells.png" class="figure-img img-fluid rounded" height="600px"> | ||
| + | |||
| + | </figure> | ||
| + | |||
| + | </html> | ||
| + | Figure 1. Functionality of UCP1-based Payload Protein in HEK-293T Cells. (e, f) Localization Pdp1NTD-EGFP-UCP1 in HEK-293T cells. For wide-field microscopy in e, cells were transfected with pNC088 (PCMV-Pdp1NTD-EGFP-UCP1). For confocal images in f, cells were co-transfected with MTS-mcherry and PNC088. Photos were taken 48 h post transfection, scale bar: 100μm for wide-field microscopy and 10 μm for confocal microscopy. Data are representative images of 3 independent experiments. (g) Charactrization of cellular metabolism in HEK-293T cells transfected with either pNC088 or pcDNA3.1(+). Glucose concentration in the cell culture medium was measured 48 h after transfection; data shows mean±SD, n=3 independent experiments. | ||
| + | <br> | ||
| + | ===Methods=== | ||
| + | To validate the function of Pdp1NTD-3*GGSGG-EGFP-2*GGSGG- UCP1,we transfected HEK-293T cells with pNC088 and observed the cellular localization of the fusion protein 48 hours post-transcription by widefield fluorescent microscopy and live-cell confocal imaging. We also analyzed the glucose consumption of the transfected cells by measuring the glucose levels in the culture medium. This analysis represented the level of cellular energy expenditure. | ||
| + | ===Results=== | ||
| + | Both wide-field fluorescent imaging (Figure 1e) and live-cell confocal imaging (Figure 1f) showed highly specific colocalization of Pdp1NTD-EGFP-UCP1 signal with mitochondria markers (MTS-mcherry, Figure 1f). Moreover, cells transfected with pNC088 showed a significantly higher glucose consumption compared to the control cells transfected with pcDNA3.1(+) vector (Figure 1g), suggesting a significantly improved energy consumption in these cells. | ||
Revision as of 16:19, 11 October 2023
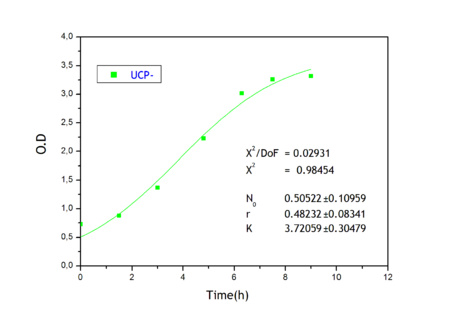
Ucp-
We used Ucp- like a negative control in our experimental design.
Ucp- has the same sequence than Ucp1 but is assembled in the opposite direction in our device.
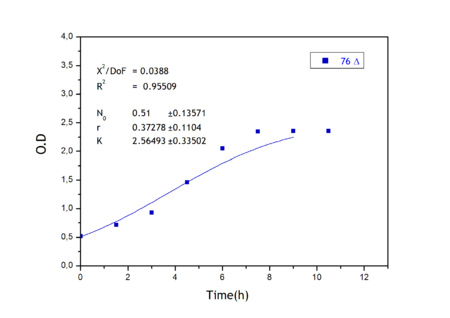
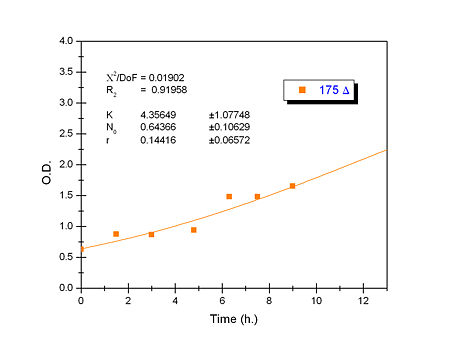
Apart from monitoring temperature evolution, we also characterized O.D. variations of each of our strains. We took O.D. measures every one a half hours for nine hours. We carried out this experiment both in Erlenmeyer flasks in the 30ºC shaking stove and in our LCCs.
This measurements were useful in order to determine some parameters for our Black Box Model. Besides, we were able to prove that the UCP was indeed being produced even though we could not see the temperature increase. Since our mutant strains Gly175Δ and Gly76Δ do not have the same growing rate as UCP+ when they are expressing the protein, the difference between the results in the strains showed that the reason for our lack of temperature increase was that we had not found the optimum conditions yet. This results made us keep on working until we obtained successful results.
Strains growth equations:
The black box model simulates the temperature evolution of the system as a function of the growth rate, galactose concentration and thermogenin expresion. We assume that our system can be reproduced by the following system of equations:
where:
- first equation: Growth of the culture: logistic growth of the different mutants used in our experiments.
- second equation: Galactose evolution: Galactose is the metabolite which induces the thermogenin expresion.
- third equation: Thermogenin concentration level.
- Fourth equation: Temperature evolution. the first term of the equation represents the losses to the ambient of the calorimeter and the second one the temperature increase as a consequence of the thermogenin expresion.
From a sample of 10 experiments in which the evolution of the temperature of the four strain was measured we perform a fit using the software simulink.
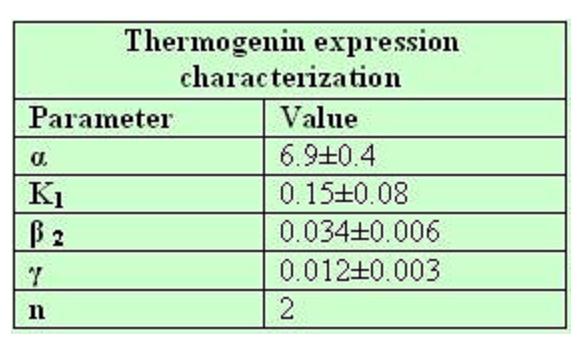
Common parameters values
Own parameters of Ucp 76 deleted activity
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal XbaI site found at 573
Illegal XbaI site found at 873
Illegal PstI site found at 289 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 289
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal XbaI site found at 573
Illegal XbaI site found at 873
Illegal PstI site found at 289 - 25INCOMPATIBLE WITH RFC[25]Illegal XbaI site found at 573
Illegal XbaI site found at 873
Illegal PstI site found at 289
Illegal AgeI site found at 726 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 674
Contribution: NUDT_CHINA 2023
In our project for iGEM2023, we aims to design a system that can effectively deliver the uncoupling protein UCP1 into adipocytes via PVCs.Herein,one composite part from the registry was used to generate the plasmid for functionality of EGFP-UCP1 Payload Protein in HEK-293T Cells .Remarkably,this part,uncoupling protein UCP1, was submitted by team iGEM08_Valencia.However, we found important information missing on the registry pages of this part. Therefore, we made efforts to provide experimental validation data and our protocol to enable the PVC-based delivery of the UCP1 part into target cells.

Methods
To validate the function of Pdp1NTD-3*GGSGG-EGFP-2*GGSGG- UCP1,we transfected HEK-293T cells with pNC088 and observed the cellular localization of the fusion protein 48 hours post-transcription by widefield fluorescent microscopy and live-cell confocal imaging. We also analyzed the glucose consumption of the transfected cells by measuring the glucose levels in the culture medium. This analysis represented the level of cellular energy expenditure.
Results
Both wide-field fluorescent imaging (Figure 1e) and live-cell confocal imaging (Figure 1f) showed highly specific colocalization of Pdp1NTD-EGFP-UCP1 signal with mitochondria markers (MTS-mcherry, Figure 1f). Moreover, cells transfected with pNC088 showed a significantly higher glucose consumption compared to the control cells transfected with pcDNA3.1(+) vector (Figure 1g), suggesting a significantly improved energy consumption in these cells.