Difference between revisions of "Part:BBa R0061"
Smelissali (Talk | contribs) |
(→Modification and Applications by BS_United_China 2023) |
||
| (33 intermediate revisions by 7 users not shown) | |||
| Line 19: | Line 19: | ||
<partinfo>BBa_R0061 parameters</partinfo> | <partinfo>BBa_R0061 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | ===Contribution=== | ||
| + | Group: Valencia_UPV iGEM 2018 | ||
| + | <br> | ||
| + | Author: Adrián Requena Gutiérrez, Carolina Ropero, Carlos Andreu Vilarroig | ||
| + | <br> | ||
| + | Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method | ||
| + | <br> | ||
| + | Documentation: | ||
| + | In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection part collection] of parts compatible with the Golden Gate assembly method, we made the part [https://parts.igem.org/Part:BBa_K2656002 BBa_K2656002] which is this part adapted to the Golden Gate technology. | ||
| + | |||
| + | In order characterize this promoter we constructed the composite part [https://parts.igem.org/Part:BBa_K2656116 BBa_K2656116] using the following RBS, CDS and terminator: | ||
| + | <html> | ||
| + | <ul> | ||
| + | <li></html>RBS [https://parts.igem.org/Part:BBa_K2656009 BBa_K2656009]: the [https://parts.igem.org/Part:B0030 B0030] RBS in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]<html></li> | ||
| + | <li></html>CDS [https://parts.igem.org/Part:BBa_K2656022 BBa_K2656022]: the [https://parts.igem.org/Part:E0040 E0040] GFPmut3b sequence in its Golden Braid standardized version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]<html></li> | ||
| + | <li></html>Terminator [https://parts.igem.org/Part:BBa_K2656026 BBa_K2656026]: the [https://parts.igem.org/Part:BBa_B0015 B0015] transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]<html></li> | ||
| + | </ul> | ||
| + | </html> | ||
| + | |||
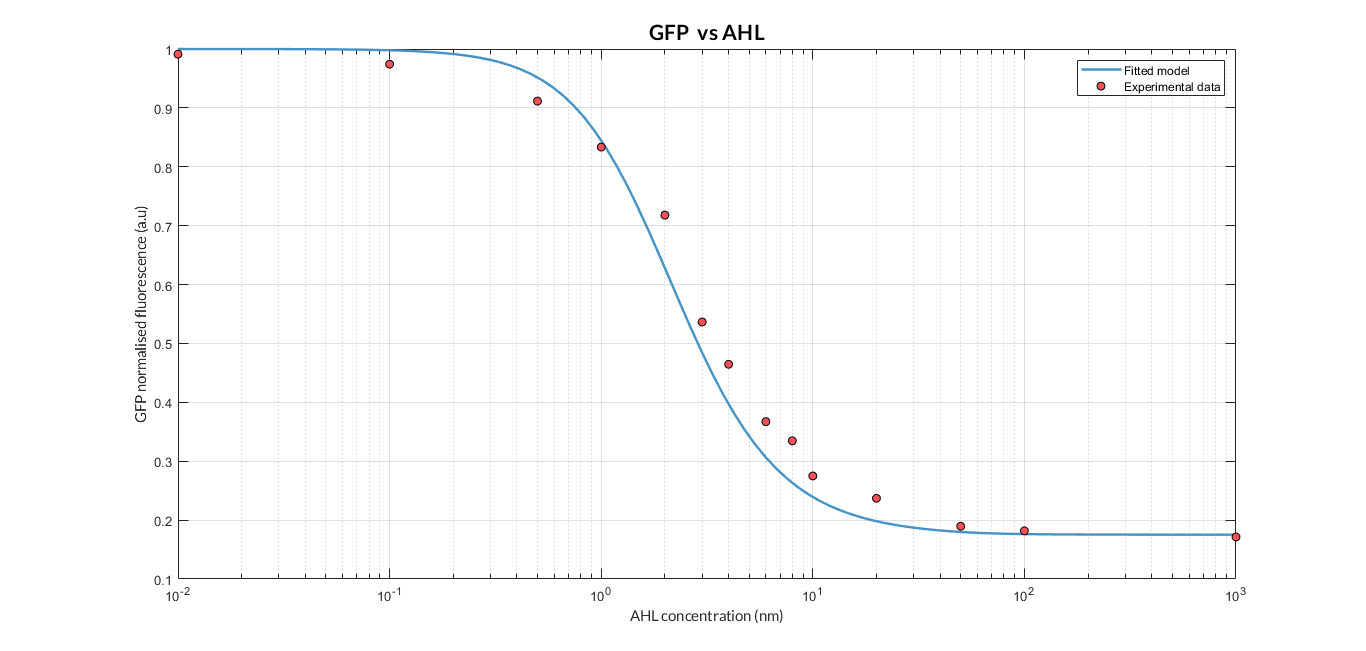
| + | By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol] and inducing at different AHL concentrations, we have obtained the following experimental measures (in blue dots). These measurements were compared with our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive AHL-LuxR model] to test it. | ||
| + | |||
| + | [[File:T--Valencia_UPV_AHL_UPV2018.png|900px|thumb|none|alt=AHL experiment.|Figure 3. AHL induction experiment and comparison with our model simulation.]] | ||
| + | |||
| + | ==<b>Characterization by BS_United_China 2021</b>== | ||
| + | See detail in part <partinfo>BBa_K3882001</partinfo> and <partinfo>BBa_K3882000</partinfo> | ||
| + | In our design, we also used LuxR regulatory protein to identify AHL in the environment. Through a series of experiments, we proved that LuxR protein and LuxR operon can correctly activate downstream genes.The downstream PVDQ-eGFP protein activated by the LuxR operon smoothly initiates transcription. We use the catabolite activator protein binding site to enhance the Luxr operator and promote the expression of down-stream genes inculding mCherry and PVDQ-GFP. | ||
| + | |||
| + | To put EL222 binding region into LUXI promoter, we need to remove the..... | ||
| + | |||
| + | <br/><li style="display: inline-block;"> [[File:Contribute08IPTG.jpg|thumb|center|500px|'''Figure1: The mCherry is expressed when <i>E. bsuahlscout</i> sensing the AHL in the environment.]] | ||
| + | <!-- --> | ||
| + | |||
| + | The LB media which contains AHL released from the <i>P. aeruginosa</i> can active the E. bsuahlscout and show visible red in the bottles (Figure 1). | ||
| + | |||
| + | <br/><li style="display: inline-block;"> [[File:K2000.png|thumb|center|800px|'''Figure 2: the standard curve of sensing the AHL in <i>E. bsuahlscout</i>.]] | ||
| + | <!-- --> | ||
| + | |||
| + | Base on the new activator binding site and LuxR operator, our <i>E. bsuahlscout</i> can be more sensitive to the low AHL in the environment (Figure 2). | ||
| + | |||
| + | |||
| + | ==Modification and Applications by BS_United_China 2023== | ||
| + | BS_United_China 2023: we utilized this part to create our blue light suicide system, which is a system that will start when our engineering bacteria lost in the external environment is not exposed to blue light. This region of luxI promoter will be used in our design, where the EL222 binding region is located between −35 (TTGACA) and −10 (TATAAT) regions of the luxI promoter. We kept the -35 and -10 region of this BBa_R0061, but changed the middle part of this luxI promoter, where we replaced it with the EL222 binding region. As EL222 is able to homodimerize under blue light, during normal laboratory or under daytime conditions, it would induce conformational change in its LOV and HTH domains to form dimers which would later bind to the EL222 binding regions. Therefore, this binding causes the release of the RNA polymerase (RNAP) sitting also in between the -35 to -10 luxI promoter region. With this modification of this part, our team is able to use the LuxI promoter in combination with EL222 protein to "switch on" the CAS9 system downstream, in order to achieve our goal of killing the bacteria. | ||
| + | |||
| + | In this way, once blue light is undetectable by EL222, the formation of EL222 dimers is terminated and the RNAP is allowed to bind back to the -35 to -10 region of the luxI promoter. This thereby activates the expression of CAS9 and its sgRNA sequence. The gene editing process takes place after that, disrupting two of the most crucial parts of the bacterial cell’s metabolic pathways, energy and protein production, which eventually cause the death of bacteria. | ||
| + | |||
| + | The detailed design is showcased below: | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.wiki/teams/4897/wiki/safety/fig-10-off-state-blue-light-suircide-system.png" width="500" height="auto">/ | ||
| + | </html> | ||
| + | |||
| + | <html> | ||
| + | <img src="https://static.igem.wiki/teams/4897/wiki/safety/fig-11-on-state-blue-light-suicide-system.png" width="500" height="auto">/ | ||
| + | </html> | ||
Latest revision as of 02:41, 10 October 2023
Promoter (HSL-mediated luxR repressor)
This part involves the -10 binding site, the -35 binding site, and the twenty nucleotides between that constitute the lux box. With this part, LuxR functions as a acyl-homoserine lactone-dependent repressor. LuxR resonds to the HSL produced by LuxI, N-(3-oxohexanoyl)-HSL. The Lux box is positioned such that it partially overlaps the consensus -35 and -10 hexamers of an RNA polymerase binding site.
Usage and Biology
A quorum-sensing system involving LuxR, the transcriptional activator, and an acyl-homserine lactone signal regulate the lux operon in vibrio fischeri. In [http://en.wikipedia.org/wiki/Vibrio_fischeri vibrio fischeri], the lux box, which is a 20-base inverted repeat unit, is positioned 42.5 bases upstream of the transcriptional start of the lux operon and is required for transcriptional activation. Egland and Greenberg constructed an artificial lacZ promoter with the lux box positioned between and overlapping the -35 and -10 hexamers of the RNA polymerase binding site was constructed, and LuxR functioned as an acyl-homoserine lactone-dependent repressor at this promoter. It was found that full-length LuxR by itself can bind to lux box-containing DNA. (see reference below)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero, Carlos Andreu Vilarroig
Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation:
In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection part collection] of parts compatible with the Golden Gate assembly method, we made the part BBa_K2656002 which is this part adapted to the Golden Gate technology.
In order characterize this promoter we constructed the composite part BBa_K2656116 using the following RBS, CDS and terminator:
- RBS BBa_K2656009: the B0030 RBS in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- CDS BBa_K2656022: the E0040 GFPmut3b sequence in its Golden Braid standardized version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
- Terminator BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol] and inducing at different AHL concentrations, we have obtained the following experimental measures (in blue dots). These measurements were compared with our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive AHL-LuxR model] to test it.
Characterization by BS_United_China 2021
See detail in part BBa_K3882001 and BBa_K3882000 In our design, we also used LuxR regulatory protein to identify AHL in the environment. Through a series of experiments, we proved that LuxR protein and LuxR operon can correctly activate downstream genes.The downstream PVDQ-eGFP protein activated by the LuxR operon smoothly initiates transcription. We use the catabolite activator protein binding site to enhance the Luxr operator and promote the expression of down-stream genes inculding mCherry and PVDQ-GFP.
To put EL222 binding region into LUXI promoter, we need to remove the.....
The LB media which contains AHL released from the P. aeruginosa can active the E. bsuahlscout and show visible red in the bottles (Figure 1).
Base on the new activator binding site and LuxR operator, our E. bsuahlscout can be more sensitive to the low AHL in the environment (Figure 2).
Modification and Applications by BS_United_China 2023
BS_United_China 2023: we utilized this part to create our blue light suicide system, which is a system that will start when our engineering bacteria lost in the external environment is not exposed to blue light. This region of luxI promoter will be used in our design, where the EL222 binding region is located between −35 (TTGACA) and −10 (TATAAT) regions of the luxI promoter. We kept the -35 and -10 region of this BBa_R0061, but changed the middle part of this luxI promoter, where we replaced it with the EL222 binding region. As EL222 is able to homodimerize under blue light, during normal laboratory or under daytime conditions, it would induce conformational change in its LOV and HTH domains to form dimers which would later bind to the EL222 binding regions. Therefore, this binding causes the release of the RNA polymerase (RNAP) sitting also in between the -35 to -10 luxI promoter region. With this modification of this part, our team is able to use the LuxI promoter in combination with EL222 protein to "switch on" the CAS9 system downstream, in order to achieve our goal of killing the bacteria.
In this way, once blue light is undetectable by EL222, the formation of EL222 dimers is terminated and the RNAP is allowed to bind back to the -35 to -10 region of the luxI promoter. This thereby activates the expression of CAS9 and its sgRNA sequence. The gene editing process takes place after that, disrupting two of the most crucial parts of the bacterial cell’s metabolic pathways, energy and protein production, which eventually cause the death of bacteria.
The detailed design is showcased below:
 /
/
 /
/