Difference between revisions of "Part:BBa K4438204"
Neha adarsh (Talk | contribs) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 8: | Line 8: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | Inspired by the fluorescence property of the DFHBI bound Spinach2.1 aptamer complex, we aimed to enhance the detection limits of this aptamer by modifying its sequence. To check whether the proposed modifications would enhance the fluorescence intensity, we performed in-silico studies by simulating the native DFHBI-Spinach aptamer complex (PDB entry: 4TS2) to validate the forcefield and other parameters to run Umbrella Sampling MD in Gromacs. Later, the modified Spinach structure was generated in X3DNA where the base mutations, A12G and U86C, are incorporated in the 4TS2 to give the DFHBI bound Spinach aptamer structure, Spinach2.6. Both the native and modified Spinach structures were subjected to identical simulation parameters in order to estimate the binding energy (ΔGbind) using the potential of mean force (PMF). The PMF was extracted from a series of umbrella sampling simulations are anticipated to provide insights into the affinity of DFHBI binding to the G-quadruplex, a part of the Spinach structure. | ||
| + | |||
| + | [[File:Parts-mod-Figure1_(1).jpg]] | ||
| + | |||
| + | <h3>GROMACS simulations</h3> | ||
| + | |||
| + | <p>The input file for both the structures were generated using CHARMM-GUI, where the RNA aptamer and ligand (DFHBI) complex reside in the water (TIP3P water system) box consisting of the defined periodic boundaries. The Amber99-OL3 RNA force field was used to perform the energy minimization and equilibration (frame rate, 125000 ns steps x 2 fs) by normal volume and temperature (NVT) method for 250 ps to get the equilibrated structures of both the native and modified RNA aptamers. Steer MD simulations were performed on these equilibrated complex structures where an imaginary spring force was used to pull the ligand out of the binding site. The RNA was restrained in the course of pulling the ligand out of their trajectories where the ligand coming off from the G-quadruplex is plotted. Later a window spacing was performed to generate configurations to help visualize the unbinding and rebinding followed by adding-up the configuration free energies to obtain the final histogram and the total binding energy.</p> | ||
| + | |||
| + | [[File:Parts-mod-Figure4_(4).jpg]] | ||
| + | |||
| + | <p>The energy minimization results were plotted using the steepest descent algorithm and it was observed that the native structure converged in almost 200 steps while for Spinach2.6 converged in nearly 180 steps. An NVT equilibration was performed and trajectory files were generated to visualise the simulation. It was found that the system reached equilibrium at less than 20 ps. This equilibrated system was subjected to the Steer MD simulations to dislocate the ligand from the G-Quadruplex binding site with spring force 10000 KJ/mol/nm2. The force applied was able to pull the ligand apart from the binding site of the RNA aptamer as seen in the animation below. Overall, the energy minimization plot suggests that the Spinach2.6 complex structure is relatively stable when compared with the native structure. Moreover, the Steer MD plots for both these complexes are significantly different suggesting the binding affinities of the ligand to RNA aptamer structure are different. There needs to perform additional simulations and experimental confirmations to strengthen this observation.</p> | ||
<!-- --> | <!-- --> | ||
Latest revision as of 01:15, 14 October 2022
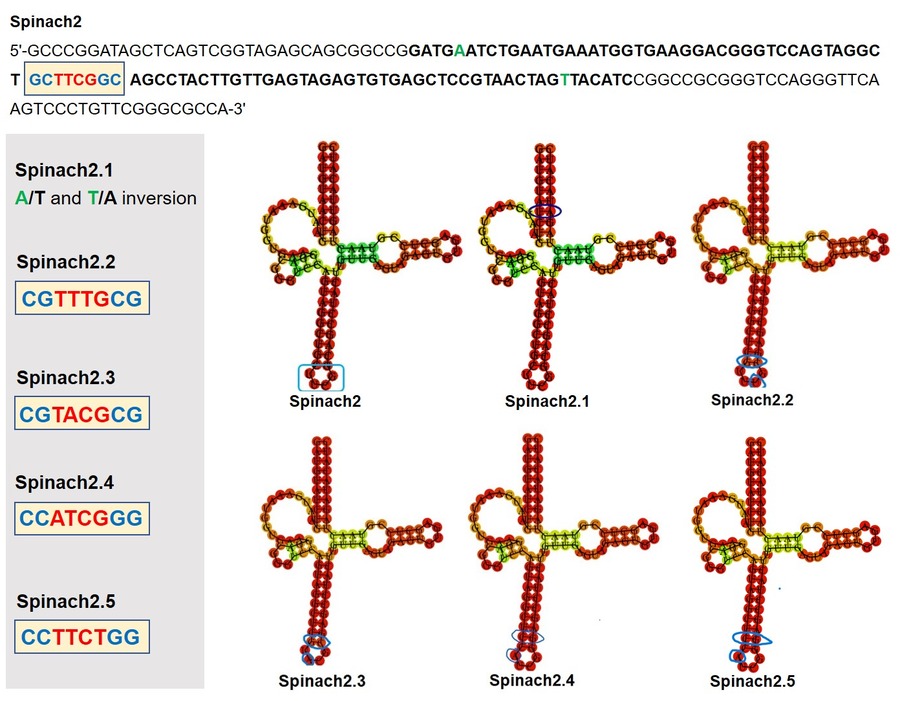
Spinach2.6
Spinach 2.6 is a mutant version of Spinach 2.1(BBa_K1330000), made to increase the fluorescent intensity given out by the aptamer on binding with DFHBI.
Usage and Biology
Inspired by the fluorescence property of the DFHBI bound Spinach2.1 aptamer complex, we aimed to enhance the detection limits of this aptamer by modifying its sequence. To check whether the proposed modifications would enhance the fluorescence intensity, we performed in-silico studies by simulating the native DFHBI-Spinach aptamer complex (PDB entry: 4TS2) to validate the forcefield and other parameters to run Umbrella Sampling MD in Gromacs. Later, the modified Spinach structure was generated in X3DNA where the base mutations, A12G and U86C, are incorporated in the 4TS2 to give the DFHBI bound Spinach aptamer structure, Spinach2.6. Both the native and modified Spinach structures were subjected to identical simulation parameters in order to estimate the binding energy (ΔGbind) using the potential of mean force (PMF). The PMF was extracted from a series of umbrella sampling simulations are anticipated to provide insights into the affinity of DFHBI binding to the G-quadruplex, a part of the Spinach structure.
GROMACS simulations
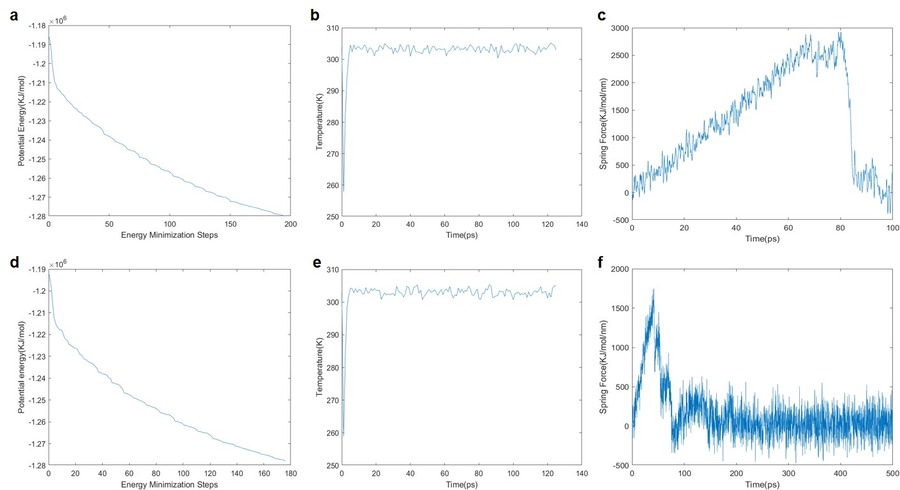
The input file for both the structures were generated using CHARMM-GUI, where the RNA aptamer and ligand (DFHBI) complex reside in the water (TIP3P water system) box consisting of the defined periodic boundaries. The Amber99-OL3 RNA force field was used to perform the energy minimization and equilibration (frame rate, 125000 ns steps x 2 fs) by normal volume and temperature (NVT) method for 250 ps to get the equilibrated structures of both the native and modified RNA aptamers. Steer MD simulations were performed on these equilibrated complex structures where an imaginary spring force was used to pull the ligand out of the binding site. The RNA was restrained in the course of pulling the ligand out of their trajectories where the ligand coming off from the G-quadruplex is plotted. Later a window spacing was performed to generate configurations to help visualize the unbinding and rebinding followed by adding-up the configuration free energies to obtain the final histogram and the total binding energy.
The energy minimization results were plotted using the steepest descent algorithm and it was observed that the native structure converged in almost 200 steps while for Spinach2.6 converged in nearly 180 steps. An NVT equilibration was performed and trajectory files were generated to visualise the simulation. It was found that the system reached equilibrium at less than 20 ps. This equilibrated system was subjected to the Steer MD simulations to dislocate the ligand from the G-Quadruplex binding site with spring force 10000 KJ/mol/nm2. The force applied was able to pull the ligand apart from the binding site of the RNA aptamer as seen in the animation below. Overall, the energy minimization plot suggests that the Spinach2.6 complex structure is relatively stable when compared with the native structure. Moreover, the Steer MD plots for both these complexes are significantly different suggesting the binding affinities of the ligand to RNA aptamer structure are different. There needs to perform additional simulations and experimental confirmations to strengthen this observation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 36
Illegal NgoMIV site found at 48
Illegal AgeI site found at 80 - 1000COMPATIBLE WITH RFC[1000]