Difference between revisions of "Part:pSB1K04"
Svetlana ia (Talk | contribs) (→Uppsala 2020's Addition) |
|||
| (4 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
pOdd4 Loop Vector based on pSB1K3 | pOdd4 Loop Vector based on pSB1K3 | ||
| + | |||
| + | |||
| + | {{CSS/DNA_Table}} | ||
| + | {{TypeIIS/Plasmids/Level1}} | ||
| + | |||
| + | |||
| + | {{TypeIIS/References}} | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 25: | ||
<partinfo>pSB1K04 parameters</partinfo> | <partinfo>pSB1K04 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ==Uppsala 2020's Addition== | ||
| + | <html> | ||
| + | <p>Since high copy number plasmids are not suitable for higher level assemblies, we have assembled 3 sets of new pOdd plasmids for different situations:</p> | ||
| + | |||
| + | <ul> | ||
| + | <li>pSB3K0# (</html><partinfo>BBa_K3425005</partinfo><html> to </html><partinfo>BBa_K3425008</partinfo><html>): medium copy number (10-12 copies)</li> | ||
| + | <li>pSB4K0# (</html><partinfo>BBa_K3425009</partinfo><html> to </html><partinfo>BBa_K3425012</partinfo><html>): low copy number (~5 copies) </li> | ||
| + | <li>pSB4A0# (</html><partinfo>BBa_K3425013</partinfo><html> to </html><partinfo>BBa_K3425016</partinfo><html>): low copy number (~5 copies) with different resistance for cotransformations</li> | ||
| + | </ul> | ||
| + | </html> | ||
| + | |||
| + | ==TU_Dresden 2022's Addition== | ||
| + | To make a plasmid suitable for yeast, we made several modification to pOdd vector, as well as added auxotrophy selective markers: | ||
| + | *URA3 marker [https://parts.igem.org/Part:BBa_K4365022 BBa_K4365022] | ||
| + | *HIS3 marker [https://parts.igem.org/Part:BBa_K4365024 BBa_K4365024] | ||
| + | *TRP1 marker [https://parts.igem.org/Part:BBa_K4365023 BBa_K4365023] | ||
Latest revision as of 20:46, 13 October 2022
pOdd4 Loop Vector based on pSB1K3
pOdd4 Loop Vector based on pSB1K3
Assembling into Level 1 Plasmds
pSB1K01-pSB1K04 series of plasmids are based on the pOdd set of Loop Vectors and are built upon the pSB1K3 plasmid backbone.
Note:
- Since pSB1K3 is a high-copy plasmid backbone, these vectors should not be used for Level 3 assemblies.
- Currently, Registry samples of these plasmid backbones have not been fully sequenced. Only their prefix and suffix along with their insert BBa_J04454 have been verified.
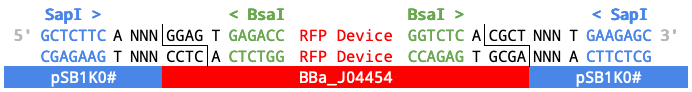
Simplified diagram of BBa_J04454 and pSB1K0# with BsaI cut site.
When four Level 0 parts are assembled into the plasmid backbone (paired with BBa_J04454) in a one-pot assembly with BsaI...

the resulting transcriptional unit (TU) will be flanked by SapI restriction sites and the specified 3 bp Fusion Sites (NNN).

Level 1 Plasmid Set
Level 1 transcriptional units in this set of plasmid backbones will be flanked by SapI recognition sites and specified 3bp fusion sites, enabling further assembly of up to 4 transcriptional units as a Level 2 Assembly.
| Registry Name | Loop Name | Fusion Site 5' | TU | Fusion Site 3' |
|---|---|---|---|---|
| pSB1K01 | pOdd1 | ATG | TU 1 | GCA |
| pSB1K02 | pOdd2 | GCA | TU 2 | TAC |
| pSB1K03 | pOdd3 | TAC | TU 3 | CAG |
| pSB1K04 | pOdd4 | CAG | TU 4 | GGT |
Level 1 -> Level 2 Assembly
Up to 4 Level 1 transcriptional units can be assembled into a Level 2 multi-transcriptional unit. By planning your assemb
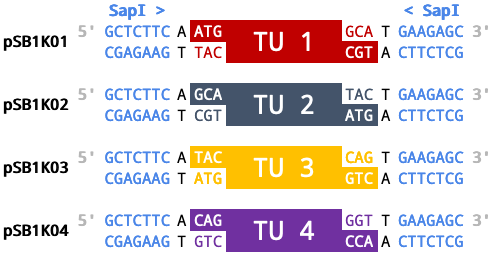
Simplified diagram of pSB1K0# set with assembled transcriptional units (TUs).
The iGEM Type IIS assembly standard is based on MoClo and Loop
- A Modular Cloning System for Standardized Assembly of Multigene Constructs
- Loop assembly: a simple and open system for recursive fabrication of DNA circuits
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2179
Illegal PstI site found at 12 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2179
Illegal PstI site found at 12 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2179
Illegal XhoI site found at 1029
Illegal XhoI site found at 2055 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2179
Illegal PstI site found at 12 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2179
Illegal PstI site found at 12 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 2185
Illegal SapI.rc site found at 5
Uppsala 2020's Addition
Since high copy number plasmids are not suitable for higher level assemblies, we have assembled 3 sets of new pOdd plasmids for different situations:
- pSB3K0# (BBa_K3425005 to BBa_K3425008): medium copy number (10-12 copies)
- pSB4K0# (BBa_K3425009 to BBa_K3425012): low copy number (~5 copies)
- pSB4A0# (BBa_K3425013 to BBa_K3425016): low copy number (~5 copies) with different resistance for cotransformations
TU_Dresden 2022's Addition
To make a plasmid suitable for yeast, we made several modification to pOdd vector, as well as added auxotrophy selective markers:
- URA3 marker BBa_K4365022
- HIS3 marker BBa_K4365024
- TRP1 marker BBa_K4365023
