Difference between revisions of "Part:BBa K4365024"
Svetlana ia (Talk | contribs) |
|||
| (One intermediate revision by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K4365024 short</partinfo> | <partinfo>BBa_K4365024 short</partinfo> | ||
| − | backbone for | + | The '''pSB1KY-HIS3''' is a shuttle plasmid backbone suitable for the iGEM '''RFC1000 assembly''' in ''Saccharomyces cerevisiae'' with ''HIS3'' auxotrophy. It was adapted from pSB1K plasmid and optimized to be expressed in ''Escherichia coli'' cells and S. cerevisiae with his3 mutations. |
| − | |||
| − | |||
| − | + | {| class="wikitable" style="margin-left: auto; margin-right: auto; border: none; width:600px;" | |
| + | |+ Collection of shuttle vector backbones optimized for ''Saccharomyces cerevisiae'' | ||
| + | |- | ||
| + | ! Backbone ID | ||
| + | ! Selection marker | ||
| + | ! Marker sequence source | ||
| + | |- style="text-align:center;" | ||
| + | |[https://parts.igem.org/wiki/index.php?title=Part:BBa_K4365022 BBa_K4365022]||''URA3''||[http://freegenes.github.io/genes/BBF10K_000474.html Open Yeast Collection ScURA3-marker] | ||
| + | |- style="text-align:center;" | ||
| + | |[https://parts.igem.org/wiki/index.php?title=Part:BBa_K4365023 BBa_K4365023]||''TRP1''||[http://freegenes.github.io/genes/BBF10K_000473.html Open Yeast Collection ScTRP1-marker] | ||
| + | |- style="text-align:center;" | ||
| + | |[https://parts.igem.org/wiki/index.php?title=Part:BBa_K4365024 BBa_K4365024]||''HIS3''||[http://freegenes.github.io/genes/BBF10K_000468.html Open Yeast Collection ScHIS3-marker] | ||
| + | |} | ||
| + | |||
| + | <br> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K4365024 SequenceAndFeatures</partinfo> | <partinfo>BBa_K4365024 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ===Usage and Biology=== | ||
| + | ''HIS3'' codes for the Imidazoleglycerol-phosphate dehydratase that catalyzes the sixth step in histidine biosynthesis <ref>https://www.yeastgenome.org/locus/S000005728</ref>. It complements his3 mutations allowing yeast to grow on media with no histidine supplements. | ||
| + | |||
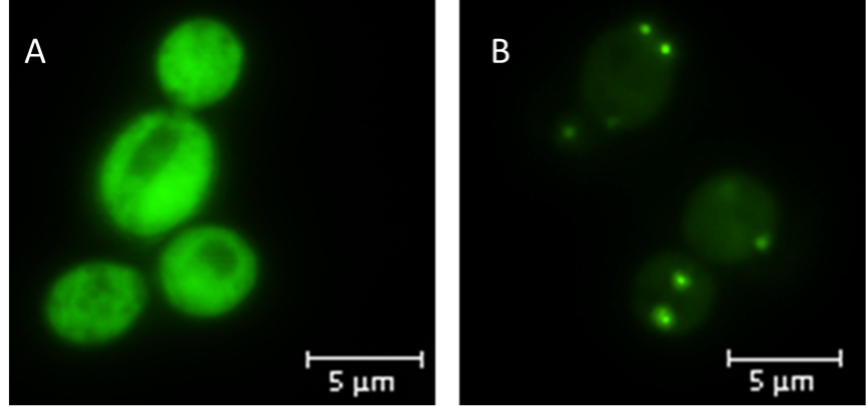
| + | The pSB1KY-HIS3 was assembled similarly to <html><a href="https://parts.igem.org/Part:BBa_K4365022#Advantages_of_pSB1KY_shuttle_vectors">pSB1KY-URA3</a></html> and was used by <html><a href="http://2022.igem.wiki/wwu-muenster/partnership">WWU-Münster 2022</a> to clone and express in ''S. cerevisiae'' the afraGFP gene and the peroxisomal targeting signal 1 (PTS1) as a fusion protein <a href="https://parts.igem.org/Part:BBa_K4188006">K4188006</a></html>. All peroxisomal proteins are translated in the cytoplasm and possess a peroxisomal targeting signal that is recognized by a receptor and can direct the protein to the peroxisome (Figure 1). | ||
| + | |||
| + | [[File:BBa_K4365023_WWU_Muenster.png|center|thumb|400px|Figure 1: fluorescence microscopy of ''S. cerevisiae'' cells with strong afraGFP signal in (A) cytosol and (B) peroxisomes. Image provided by <html><a href="http://2022.igem.wiki/wwu-muenster">WWU-Münster 2022</a></html>.]] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 08:16, 12 October 2022
pSB1KY-HIS3
The pSB1KY-HIS3 is a shuttle plasmid backbone suitable for the iGEM RFC1000 assembly in Saccharomyces cerevisiae with HIS3 auxotrophy. It was adapted from pSB1K plasmid and optimized to be expressed in Escherichia coli cells and S. cerevisiae with his3 mutations.
| Backbone ID | Selection marker | Marker sequence source |
|---|---|---|
| BBa_K4365022 | URA3 | [http://freegenes.github.io/genes/BBF10K_000474.html Open Yeast Collection ScURA3-marker] |
| BBa_K4365023 | TRP1 | [http://freegenes.github.io/genes/BBF10K_000473.html Open Yeast Collection ScTRP1-marker] |
| BBa_K4365024 | HIS3 | [http://freegenes.github.io/genes/BBF10K_000468.html Open Yeast Collection ScHIS3-marker] |
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal XbaI site found at 462
Illegal SpeI site found at 1470
Illegal PstI site found at 1 - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal SpeI site found at 1470
Illegal PstI site found at 1
Illegal NotI site found at 2594 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BamHI site found at 97
Illegal XhoI site found at 3522
Illegal XhoI site found at 4548 - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal XbaI site found at 462
Illegal SpeI site found at 1470
Illegal PstI site found at 1 - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal XbaI site found at 462
Illegal SpeI site found at 1470
Illegal PstI site found at 1 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Usage and Biology
HIS3 codes for the Imidazoleglycerol-phosphate dehydratase that catalyzes the sixth step in histidine biosynthesis [1]. It complements his3 mutations allowing yeast to grow on media with no histidine supplements.
The pSB1KY-HIS3 was assembled similarly to pSB1KY-URA3 and was used by WWU-Münster 2022 to clone and express in ''S. cerevisiae'' the afraGFP gene and the peroxisomal targeting signal 1 (PTS1) as a fusion protein K4188006. All peroxisomal proteins are translated in the cytoplasm and possess a peroxisomal targeting signal that is recognized by a receptor and can direct the protein to the peroxisome (Figure 1).