Difference between revisions of "Part:BBa K4140024"
(→Part Description) |
|||
| (13 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
| + | <partinfo>BBa_K4140024 short</partinfo> | ||
| + | ==Part Description== | ||
| + | The LacZ-alpha fragment is encoded by this region and is derived from the pUC19 cloning vector. Because it is smaller than the LacZ-omega region, the LacZ-alpha fragment can be easily incorporated into a plasmid when the two non-functional LacZ gene fragments (alpha and omega) are co-expressed. X-gal (5-bromo-4-chloro-3-indoyl-d-galactopyranoside), a soluble colorless molecule that is a substrate of ß-galactosidase and generates a blue product when cleaved, is used to detect the alpha-complementation. It's linked to KP-SP ISA secreting peptide signal at the N-terminus of the reporter protein to transmit it from intra-cellular to extra-cellular. | ||
| + | |||
| + | ==Usage== | ||
| + | In order to detect the increased levels of phenylalanine in PKU patients, we used the lacZ alpha gene which encodes beta-galactosidase, which catalyzes the cleavage of lactose to form galactose and glucose that utilizes X-gal to change the color into blue as shown in figure 1. | ||
| + | [[File:wcb.png| ]] | ||
| + | |||
| + | Figure 1. (shows the whole cell-based biosensor.) | ||
| + | ==Characterization by mathematical modeling== | ||
| + | We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (2) and graph (1). | ||
| + | |||
| + | [[File:modell1.png|Right|]] | ||
| + | <br> | ||
| + | Figure (2) represents the cascade of reactions in whole cell-based biosensor model. | ||
| + | [[File:modell11.png|Right|]] | ||
| + | <br><br><br> | ||
| + | Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase. | ||
| + | ==Experimental Characterization== | ||
| + | [[File:capture7.png|right|]] | ||
| + | <br><br><br><br><br><br><br> | ||
| + | This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 3. The running part (ordered from IDT) included KP-SP and lacZ alpha. | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K4140024 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K4140024 parameters</partinfo> | ||
| + | <!-- --> | ||
Latest revision as of 04:41, 12 October 2022
Reporter device
Part Description
The LacZ-alpha fragment is encoded by this region and is derived from the pUC19 cloning vector. Because it is smaller than the LacZ-omega region, the LacZ-alpha fragment can be easily incorporated into a plasmid when the two non-functional LacZ gene fragments (alpha and omega) are co-expressed. X-gal (5-bromo-4-chloro-3-indoyl-d-galactopyranoside), a soluble colorless molecule that is a substrate of ß-galactosidase and generates a blue product when cleaved, is used to detect the alpha-complementation. It's linked to KP-SP ISA secreting peptide signal at the N-terminus of the reporter protein to transmit it from intra-cellular to extra-cellular.
Usage
In order to detect the increased levels of phenylalanine in PKU patients, we used the lacZ alpha gene which encodes beta-galactosidase, which catalyzes the cleavage of lactose to form galactose and glucose that utilizes X-gal to change the color into blue as shown in figure 1.
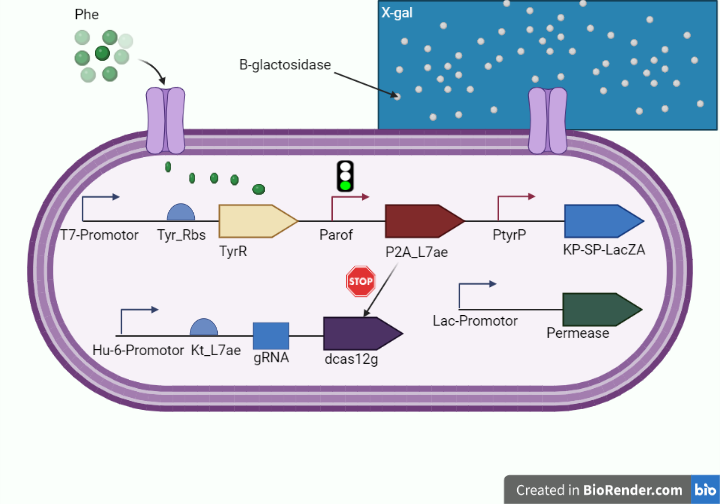
Figure 1. (shows the whole cell-based biosensor.)
Characterization by mathematical modeling
We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (2) and graph (1).
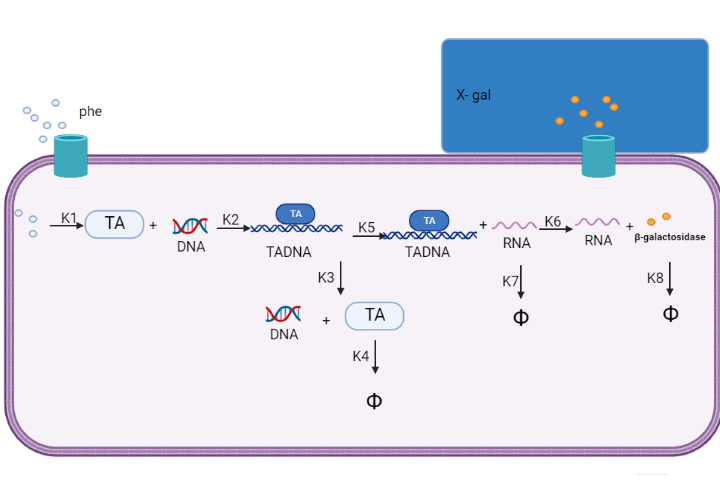
Figure (2) represents the cascade of reactions in whole cell-based biosensor model.
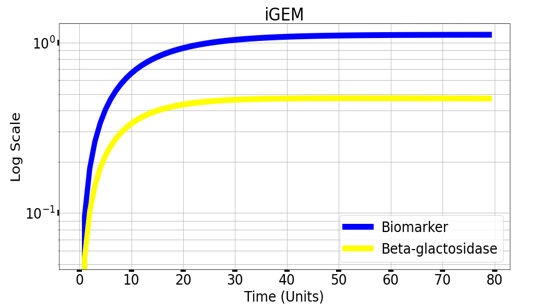
Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase.
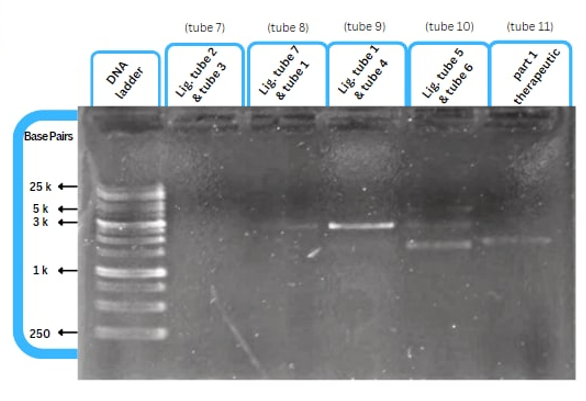
Experimental Characterization
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 3. The running part (ordered from IDT) included KP-SP and lacZ alpha.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 240
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]