Difference between revisions of "Part:BBa K4130014"
| Line 3: | Line 3: | ||
<partinfo>BBa_K4130014 short</partinfo> | <partinfo>BBa_K4130014 short</partinfo> | ||
| − | + | =Sar09-3 Aptamer= | |
| + | ssDNA Aptamer for free sarcosine with a 3’ amine modification | ||
| − | + | This part selectively detects sarcosine with a dissociation constant of 0.33 ± 0.05nM and a literature detection range of 5pM-50uM [10]. Sarcosine (N-methylglycine) is an amino acid derivative that is a byproduct of creatine hydrolysis [1] as well as an intermediate of choline metabolism [2]. In maple trees, it is found to increase in concentration from 0.01uM to 0.12uM [8] as the tree matures and nears new-leaf production (aka “bud break”), containing methyl groups that contribute an off-taste to maple syrup when boiled down from sap [3] Besides maple trees, sarcosine is also a human metabolite, thought to be a marker for prostate cancer [1], and has been investigated for use in treatment against schizophrenia. | |
| − | The | + | ===Usage and Biology=== |
| + | Aptamers are ssDNA or ssRNA molecules that have selective binding capabilities to target molecules such as proteins or small molecules. These oligonucleotides can be synthesized through PCR and selected in vitro through SELEX[4]. Their antibody-like function can be utilized to detect target molecules, particularly in diagnostic biosensor applications known as “aptasensors”[4]. However, aptamers are often easier to synthesize and therefore cheaper than antibodies, in addition to being optimal for specific targeting of smaller molecules[5] such as sarcosine. In context, this aptamer’s function was utilized in an electrochemical aptasensor mechanism, in which the aptamer was pipetted, also known as dropcasting, onto an electrode, followed by an application of current. | ||
| + | |||
| + | ===Aptamer Specifications=== | ||
| + | The initial aptamer sequence and structure for Sar09-3 (Fig. 2) was selected based on a previous study that demonstrated effective binding of sarcosine to the picomolar levels [10] After selecting the sequence, in the interest of time and effort, the ssDNA aptamer was ordered to be synthesized by IDT. In addition, the 3’ end of the aptamer sequence was ordered to be modified with an amine group (Fig. 1). This was done to ensure that the aptamers could covalently bond to functional groups present on the surface-deposited nanomaterials applied to screen printed electrodes (SPEs) to improve their performance. Strong electrode-to-aptamer binding, known as immobilization[9], is critical for establishing the accuracy and sensitivity of the aptasensor, since aptamer-to-target binding events incur folding that impedes the current of the electrode, and therefore triggers a signal in the form of a resistance readout. | ||
| + | |||
| + | [[Image:BBa_K4130000_Amination.png|500px|left|thumb|<p align="justify">'''Figure 1:''' The 3’ amination applied to S09-3. The 3’ end was chosen for modification because of literature truncation testing that, based on ΔGo values, indicated 3’ truncation did not hinder aptamer folding as much as the 5’ end[0], suggesting that 3’ modification would impose less hindrance on target binding. </p>]] | ||
| + | |||
| + | [[Image:BBa_K4130000_SarcosineAptamer1.png|500px|left|thumb|<p align="justify">'''Figure 2:''' Predicted secondary structure of Sar09-3 as modeled by the Mfold web server[0]. Highlighted in blue is the consensus sequence identified by the MEME suite, which is shown to be responsible for binding to the target molecule[6]. </p>]] | ||
| + | |||
| + | ===Characterization=== | ||
| + | The goal of utilizing aptamers was to incorporate them with electrochemical techniques in order to develop a functional aptasensor capable of testing accurate concentrations of sarcosine. | ||
| + | |||
| + | For the initial carbon SPE tests, a 0.1uM solution of the reconstituted Sar09-3 aptamer/ddH2O solution To deposit onto the electrode the solution was dropcasted (pipetted) onto the modified carbon electrode in 2uL volumes. The carbon SPEs were modified with chitosan and chemically reduced graphene oxide (rGO),To test that the aptamer was immobilized on the electrode, potentiostatic measurements via cyclic voltammetry were run between -1.0V to 1.0V at a scan rate of 0.2V/s for 10 cycles. An expected increase of resistance of about 0.1 current/uA occurred when testing the electrode after performing the aptamer immobilization protocol in comparison to the modified electrode with no deposited aptamers. This data indicates that aminating our aptamer was an effective mechanism for improving aptamer-to-electrode binding. Upon depositing and incubating the electrode in 1mM solution of the target molecule sarcosine, there was no change in resistance, suggesting that there was either not a high enough concentration of aptamer solution to produce a sensitive enough reading. | ||
| + | |||
| + | To improve sensitivity, Sar09-3 was tested with gold electrodes and electrodeposited rGO carbon electrodes, both of which were modified with chitosan and methylene blue intercalation. To saturate the surface, a 10mM rather than 0.1uM aptamer solution was dropcasted on the electrodes and differential pulse voltammetry (DPV) was run for two cycles between 0.0V - 0.8V immersed in a control PBS solution. To test the aptamer binding, the electrode was incubated in 100uL of 1M sarcosine solution for 15 minutes. The curves for both gold and carbon electrodes [Fig. 3, 4] show a significant change in resistance after contact with the sarcosine solution, demonstrating that the amine modification was compatible with multiple types of electrode modification, as well demonstrate that Sar09-3 successfully bound to the target molecule, sarcosine. Because a binding event occurred, it also implies the application of the modification did not impact the folding enough to impede Sar09-3 from binding to its target. | ||
| + | |||
| + | [[Image:BBa_K4130000_SarcosineAptamer1.png|500px|left|thumb|<p align="justify">'''Figure 3:''' The DPV curve for testing target-aptamer binding on rGO/chitosan modified carbon SPEs intercalated with methylene blue. The two test trials (orange and light blue curves) show that the aptamer binds with sarcosine, as indicated by the change in current. The conditions of the controls were as follows: red and dark blue curves show control chitosan/MB carbon SPEs, the green and purple curves show control chitosan/MB/aptamer carbon SPEs, and the curves show chitosan/aptamer gold SPEs incubated with 100uL of sarcosine. The orange and light blue peaks indicate successful binding events between Sar09-3 and sarcosine on the two separate electrodes. | ||
| + | </p>]] | ||
| + | |||
| + | [[Image:BBa_K4130000_SarcosineAptamer1.png|500px|left|thumb|<p align="justify">'''Figure 4:''' DPV curves for testing target-aptamer binding on chitosan modified gold SPEs intercalated with methylene blue. The first test trial (orange) shows that the aptamer bound to Sar09-3, as indicated by change in current. The red and dark blue curves show control chitosan modified gold SPEs, the light blue and purple curves show control chitosan-aptamer modified gold SPEs, and the orange and light blue curves show chitosan-aptamer gold SPEs not incubated with sarcosine, while the orange and green curves show chitosan-aptamer gold SPEs that were incubated with sarcosine. The failure of the second test trial (green curve) to bind to the aptamer can be attributed to human error with the crosslinking or rinsing steps regarding aptamer immobilization. </p>]] | ||
Revision as of 01:51, 12 October 2022
Aptamer for free Sarcosine
Sar09-3 Aptamer
ssDNA Aptamer for free sarcosine with a 3’ amine modification
This part selectively detects sarcosine with a dissociation constant of 0.33 ± 0.05nM and a literature detection range of 5pM-50uM [10]. Sarcosine (N-methylglycine) is an amino acid derivative that is a byproduct of creatine hydrolysis [1] as well as an intermediate of choline metabolism [2]. In maple trees, it is found to increase in concentration from 0.01uM to 0.12uM [8] as the tree matures and nears new-leaf production (aka “bud break”), containing methyl groups that contribute an off-taste to maple syrup when boiled down from sap [3] Besides maple trees, sarcosine is also a human metabolite, thought to be a marker for prostate cancer [1], and has been investigated for use in treatment against schizophrenia.
Usage and Biology
Aptamers are ssDNA or ssRNA molecules that have selective binding capabilities to target molecules such as proteins or small molecules. These oligonucleotides can be synthesized through PCR and selected in vitro through SELEX[4]. Their antibody-like function can be utilized to detect target molecules, particularly in diagnostic biosensor applications known as “aptasensors”[4]. However, aptamers are often easier to synthesize and therefore cheaper than antibodies, in addition to being optimal for specific targeting of smaller molecules[5] such as sarcosine. In context, this aptamer’s function was utilized in an electrochemical aptasensor mechanism, in which the aptamer was pipetted, also known as dropcasting, onto an electrode, followed by an application of current.
Aptamer Specifications
The initial aptamer sequence and structure for Sar09-3 (Fig. 2) was selected based on a previous study that demonstrated effective binding of sarcosine to the picomolar levels [10] After selecting the sequence, in the interest of time and effort, the ssDNA aptamer was ordered to be synthesized by IDT. In addition, the 3’ end of the aptamer sequence was ordered to be modified with an amine group (Fig. 1). This was done to ensure that the aptamers could covalently bond to functional groups present on the surface-deposited nanomaterials applied to screen printed electrodes (SPEs) to improve their performance. Strong electrode-to-aptamer binding, known as immobilization[9], is critical for establishing the accuracy and sensitivity of the aptasensor, since aptamer-to-target binding events incur folding that impedes the current of the electrode, and therefore triggers a signal in the form of a resistance readout.
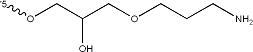
Figure 1: The 3’ amination applied to S09-3. The 3’ end was chosen for modification because of literature truncation testing that, based on ΔGo values, indicated 3’ truncation did not hinder aptamer folding as much as the 5’ end[0], suggesting that 3’ modification would impose less hindrance on target binding.
Characterization
The goal of utilizing aptamers was to incorporate them with electrochemical techniques in order to develop a functional aptasensor capable of testing accurate concentrations of sarcosine.
For the initial carbon SPE tests, a 0.1uM solution of the reconstituted Sar09-3 aptamer/ddH2O solution To deposit onto the electrode the solution was dropcasted (pipetted) onto the modified carbon electrode in 2uL volumes. The carbon SPEs were modified with chitosan and chemically reduced graphene oxide (rGO),To test that the aptamer was immobilized on the electrode, potentiostatic measurements via cyclic voltammetry were run between -1.0V to 1.0V at a scan rate of 0.2V/s for 10 cycles. An expected increase of resistance of about 0.1 current/uA occurred when testing the electrode after performing the aptamer immobilization protocol in comparison to the modified electrode with no deposited aptamers. This data indicates that aminating our aptamer was an effective mechanism for improving aptamer-to-electrode binding. Upon depositing and incubating the electrode in 1mM solution of the target molecule sarcosine, there was no change in resistance, suggesting that there was either not a high enough concentration of aptamer solution to produce a sensitive enough reading.
To improve sensitivity, Sar09-3 was tested with gold electrodes and electrodeposited rGO carbon electrodes, both of which were modified with chitosan and methylene blue intercalation. To saturate the surface, a 10mM rather than 0.1uM aptamer solution was dropcasted on the electrodes and differential pulse voltammetry (DPV) was run for two cycles between 0.0V - 0.8V immersed in a control PBS solution. To test the aptamer binding, the electrode was incubated in 100uL of 1M sarcosine solution for 15 minutes. The curves for both gold and carbon electrodes [Fig. 3, 4] show a significant change in resistance after contact with the sarcosine solution, demonstrating that the amine modification was compatible with multiple types of electrode modification, as well demonstrate that Sar09-3 successfully bound to the target molecule, sarcosine. Because a binding event occurred, it also implies the application of the modification did not impact the folding enough to impede Sar09-3 from binding to its target.
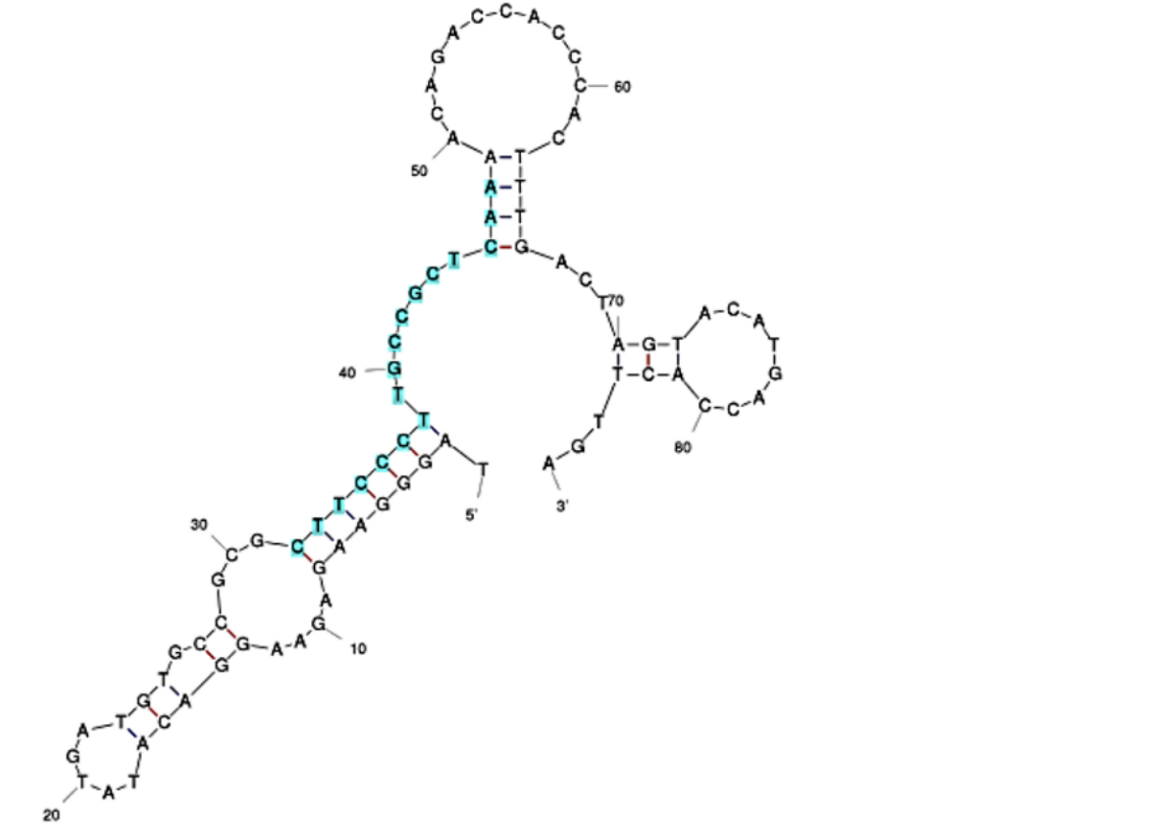
Figure 3: The DPV curve for testing target-aptamer binding on rGO/chitosan modified carbon SPEs intercalated with methylene blue. The two test trials (orange and light blue curves) show that the aptamer binds with sarcosine, as indicated by the change in current. The conditions of the controls were as follows: red and dark blue curves show control chitosan/MB carbon SPEs, the green and purple curves show control chitosan/MB/aptamer carbon SPEs, and the curves show chitosan/aptamer gold SPEs incubated with 100uL of sarcosine. The orange and light blue peaks indicate successful binding events between Sar09-3 and sarcosine on the two separate electrodes.

Figure 4: DPV curves for testing target-aptamer binding on chitosan modified gold SPEs intercalated with methylene blue. The first test trial (orange) shows that the aptamer bound to Sar09-3, as indicated by change in current. The red and dark blue curves show control chitosan modified gold SPEs, the light blue and purple curves show control chitosan-aptamer modified gold SPEs, and the orange and light blue curves show chitosan-aptamer gold SPEs not incubated with sarcosine, while the orange and green curves show chitosan-aptamer gold SPEs that were incubated with sarcosine. The failure of the second test trial (green curve) to bind to the aptamer can be attributed to human error with the crosslinking or rinsing steps regarding aptamer immobilization.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal SpeI site found at 67
- 12INCOMPATIBLE WITH RFC[12]Illegal SpeI site found at 67
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal SpeI site found at 67
- 25INCOMPATIBLE WITH RFC[25]Illegal SpeI site found at 67
- 1000COMPATIBLE WITH RFC[1000]
