Difference between revisions of "Part:BBa K1150000"
Li zhuo ran (Talk | contribs) m (adapt size of image) |
Li zhuo ran (Talk | contribs) m (adapt size of image) |
||
| Line 172: | Line 172: | ||
<p>Fig.3 Sequencing results have shown a shift mutation in target sequence</p> | <p>Fig.3 Sequencing results have shown a shift mutation in target sequence</p> | ||
| − | < img src="https://static.igem.wiki/teams/4298/wiki/proof-of-concept-box.png | + | < img src="https://static.igem.wiki/teams/4298/wiki/proof-of-concept-box.png" width="50%" /> |
| − | " width="50%" /> | + | |
<p>Fig.4 | <p>Fig.4 | ||
Revision as of 19:04, 11 October 2022
dCas9
| dCas9 | |
|---|---|
| Function | Binding protein |
| Use in | Mammalian cells |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Organism | Streptococcus pyogenes |
| Source | Feng Zhang, Addgene |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
dCas9 is a codon optimized and standardized (RFC 25) protein for human cell lines. Interacting with a DNA-binding RNA and fused with different effector domains it can be used for specific gene regulation.
Cas9 is the main protein of the CRISPR/Cas system II of Streptococcus pyogenes. CRISPR systems protect bacteria and archaea from phages by recognizing and cleaving of invading phage DNA. This recognition is based on Watson Crick base pairing between a short RNA, called crRNA, and the complementary DNA strand. A second RNA, called tracrRNA, connects crRNA and Cas9. These three parts together form a protein-RNA-DNA complex with the targeted DNA strand [1].
Cas9 became of great interest for research concerning DNA targeting, because of its ability to recognize site specific DNA strands by a crRNA.
At first the functionality of Cas9 was modified by exchanging aminoacids. As a result, Cas9 was able to introduce mutations within the genome of several organisms by causing double strand breaks [2][3]. Then, it was converted from a nuclease to a nickase introducing single strand breaks [4] and lately it was converted to an enzymatically inactive form, called dCas9 [5].
This dCas9 is codon optimized for human cell lines and standardized (RFC 25). It can be used as a DNA binding protein, that can be fused with different effectors in order to regulate gene expression.
Improvement
Characterized by Peking 2019
In our experiment, we found that the inhibitory effect of dCas9 to DNA replication shows to have over-inhibition on cell growth and small dynamic range when the sgRNAs targeting high affinity DnaA boxes. In order to extend this system to combine all DnaA boxes, we improve the performance of the CRISPRri system to weaken its effect by adding a degradation signal peptide ssrA to dCas9. Thus, we get an improved part (BBa_K3081055) from dCas9 (BBa_K1150000). The ssrA peptide tag from Mycoplasma florum has been developed as a versatile biotechnology tool to control orthogonal degradation of tagged proteins in Escherichia coli [1]. Ideally, after fused dCas9 and ssrA, it accelerates the degradation rate of dCas9 and thus weaken the inhibition of DNA replication by CRISPRri. As a preliminary verification, firstly, we fused ssrA tag to dCas9-sfGFP, we tested the expression curve of dCas9-sfGFP-ssrA by adding different concentration of inducer (Figure 1A). We found that the protein with ssrA was expressed at a lower level than the control protein, with the same concentration of inducer. We believe that because ssrA mediates protein degradation, the equilibrium concentration of the protein is reduced. To test the function of fused version of dCas9-ssrA, we co-transformed two plasmids to one E. coli cell, which express mRFP and dCas9-ssrA with sgRNA targeting CDS of mRFP, respectively. As a matter of fact, the new CRISPRi system shows a gentler decrease in fluorescence when dCas9 is fused with ssrA tag, while non-binding dCas9 with or without ssrA has no influence on mRFP expression (Figure 1B). This result confirmed fusion with ssrA, the original function of dCas9 is unaffected. So, we tested the improved CRISPRri-ssrA system with targeting site of DnaA boxes which are shown to have excessive inhibition on cell growth before. We found that the degradation tag makes inhibition effect much milder, which allows for a wider adjusting range (Figure 1C).
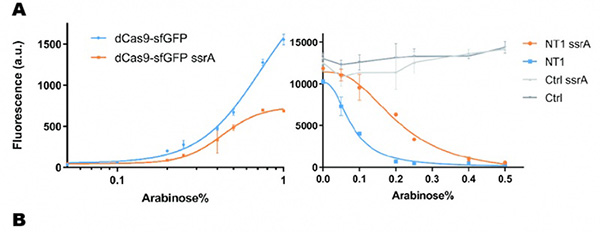


Figure 1. Characterization of CRISPRri-ssrA system. (A) Comparison between dCas9 and dCas9-ssrA system by expression level and CRISPRi effect on mRFP fluorescence. (B) Comparison of effect on cell growth between CRISPRri and CRISPRri-ssrA, both targeted to R1+ box(left:R1+; right:R1+ ssrA). (C) Reversibility of CRISPRri-ssrA system targeted to R1+ box. Hollow arrows stand for removal of arabinose while solid black arrows stand for addition of arabinose.
Reference:
[1] Lv, L., Wu, Y., Zhao, G., & Qi, H. (2019). Improvement in the Orthogonal Protein Degradation in Escherichia coli by Truncated mf-ssrA Tag. Transactions of Tianjin University, 1-7.
Usage example
designed by Peking 2020
David Liu’s lab created the first base editor in 2016 (Komor et al., 2016) and since then has been trying to expand their precision editing capabilities. Base editors make specific DNA base changes and consist of a catalytically impaired Cas protein (dCas or Cas nickase) fused to a DNA-modifying enzyme, in this case a deaminase. Base changes from C•G-to-T•A are mediated by cytosine base editors (CBEs) and base changes from A•T-to-G•C are mediated by adenine base editors (ABEs). How does this work? Through molecular biology teamwork. The guide RNA (gRNA) specifies the editing target site on the DNA, the Cas domain directs the modifying enzyme to the target site, and the deaminase induces the DNA base change without a DNA double-strand break. But base editors aren’t perfect. They may be slow, can only target certain sites, or make only a subset of base substitutions. (addgene blog by Susanna Bachle)
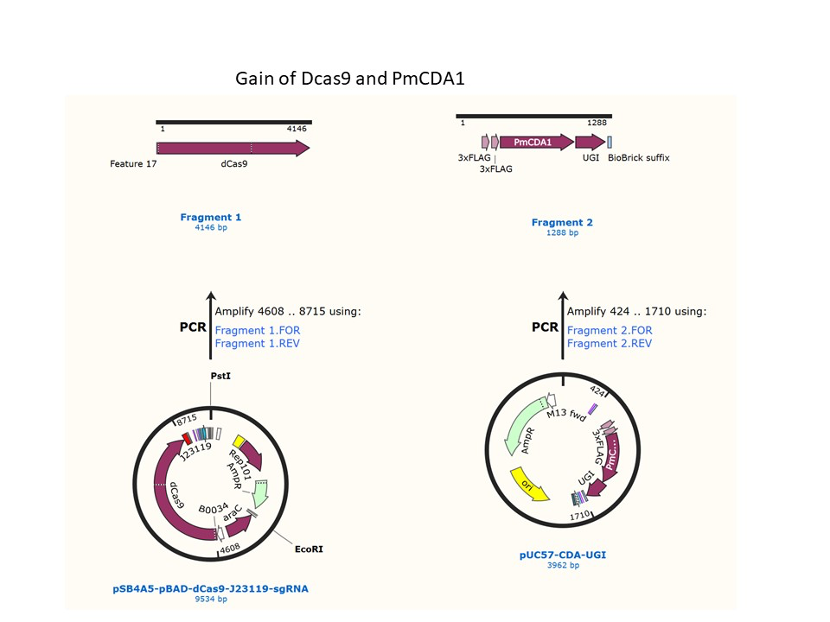
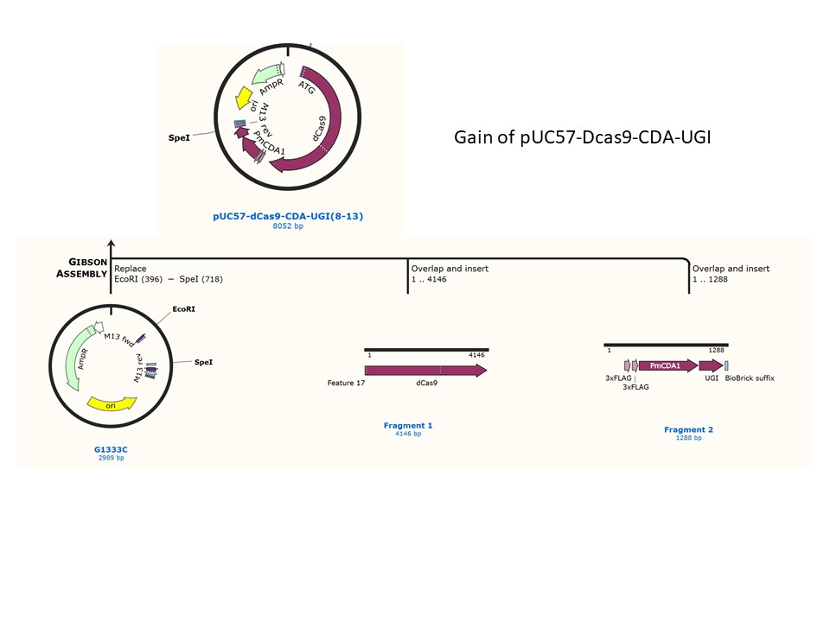
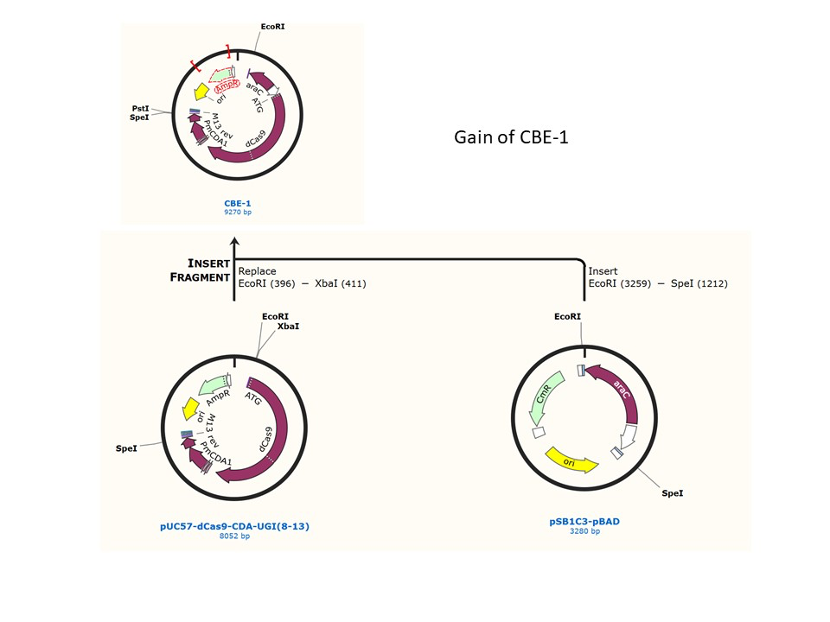
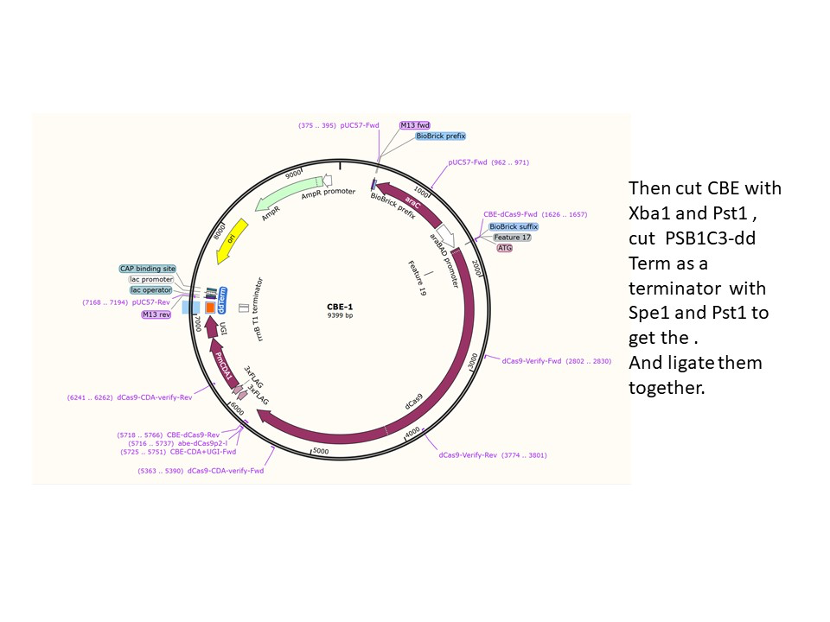
We used the existing plasmids for enzyme digestion and ligation, and ePCR was added to the BioBrick connector. After multiple rounds of splicing and assembly, we obtained the ABE and CBE we needed. The schematic diagrams are as follows:
CBE
Until 2016, precise single base changes were only possible through exploiting the homology-directed repair (HDR) pathway which occurs in cells at low frequencies and relies on the efficient delivery of donor DNA to facilitate repair. Since the development of CRISPR-mediated base editing (BE), these types of repairs can now be done more efficiently than before. A base editor precisely changes a single base with an efficiency typically ranging from 2575%, while the success of precise change via HDR limited to 0-5%. This blog post covers a brief review of different basic BE technologies and their adaptation for plant genome editing. (addgene blog by Guest Blogger)
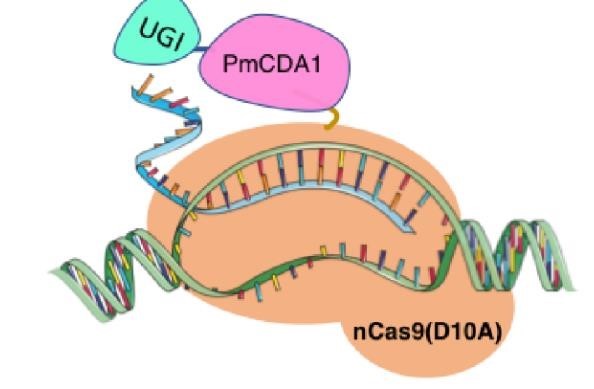
In 2016, two independent groups (komor et al., 2016 and Nishida et al., 2016) invented CRISPR base editor by linking cytosine deaminase with cas9 cleavage enzyme (ncas9), thus achieving accurate and efficient base rewriting in the genome. Ncas9 creates a gap in DNA by cutting only one single strand, thus greatly reducing the possibility of harmful insertion deletion. After binding with DNA, CBE deamination of target cytosine (C) into uracil (U) base. Later, the resulting U•G pairs were repaired through the cell mismatch repair mechanism to convert the original C•G pair into T•A, or reduced to the original C•G through the uracil glycosylase mediated base excision repair. The presence of UGI minimizes the second result and increases the production of required T•A base pairs.
CBE
Until 2016, precise single base changes were only possible through exploiting the homology-directed repair (HDR) pathway which occurs in cells at low frequencies and relies on the efficient delivery of donor DNA to facilitate repair. Since the development of CRISPR-mediated base editing (BE), these types of repairs can now be done more efficiently than before. A base editor precisely changes a single base with an efficiency typically ranging from 2575%, while the success of precise change via HDR limited to 0-5%. This blog post covers a brief review of different basic BE technologies and their adaptation for plant genome editing. (addgene blog by Guest Blogger)
In 2016, two independent groups (komor et al., 2016 and Nishida et al., 2016) invented CRISPR base editor by linking cytosine deaminase with cas9 cleavage enzyme (ncas9), thus achieving accurate and efficient base rewriting in the genome. Ncas9 creates a gap in DNA by cutting only one single strand, thus greatly reducing the possibility of harmful insertion deletion. After binding with DNA, CBE deamination of target cytosine (C) into uracil (U) base. Later, the resulting U•G pairs were repaired through the cell mismatch repair mechanism to convert the original C•G pair into T•A, or reduced to the original C•G through the uracil glycosylase mediated base excision repair. The presence of UGI minimizes the second result and increases the production of required T•A base pairs.
(addgene blog by Guest Blogger)
ABE
(Gaudelli et al., 2020.)
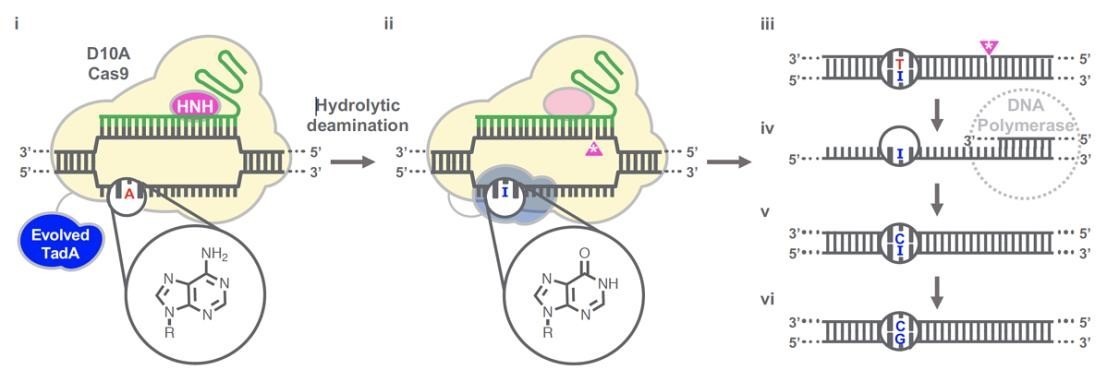
Gaudelli et al. have successfully developed an adenosine deaminase, which can act on DNA for adenine base editing. They first created a defective chloramphenicol resistance gene (CamR) by introducing a point mutation (H193Y). Reversal of this mutation by adenine base editor will restore antibiotic resistance. To find such a protein, they created a mutant library of E.coli tRNA adenosine deaminase (ecTadA), fused it with dcas9, and transformed it into E.coli containing the defective CamR gene. Screening of viable colonies and subsequent rounds of evolution and engineering produced a mutant TadA (TadA *), which accepted DNA as a substrate satisfactorily.
The artificially evolved adenosine deaminase catalyzes the transformation of target "A" into "I" (inosine), which is regarded as "G" by cell polymerase. Subsequently, a primitive genome A•T base pair was transformed into a G•C base pair. Since inosine excision repair is not as active as uracil excision, ABE does not require any additional inhibitor proteins, such as UGI in CBE.
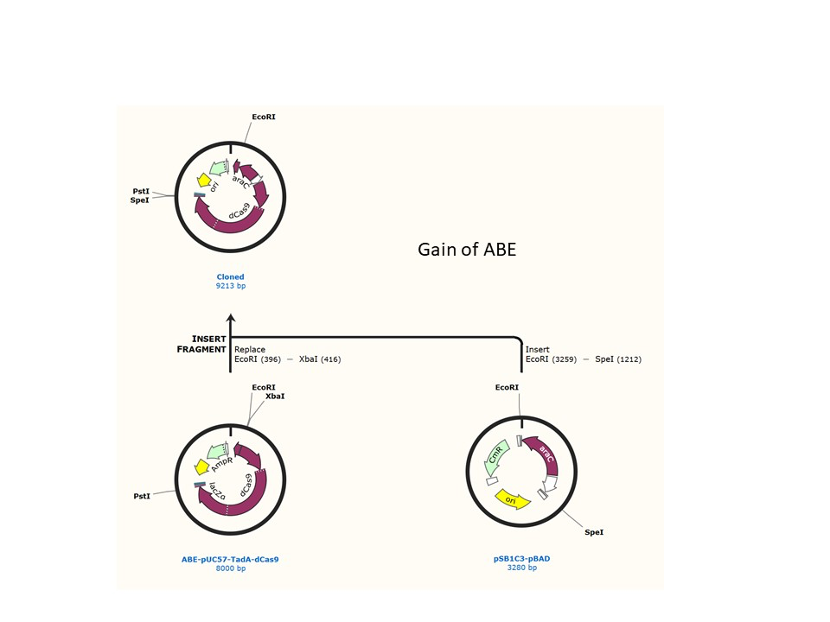
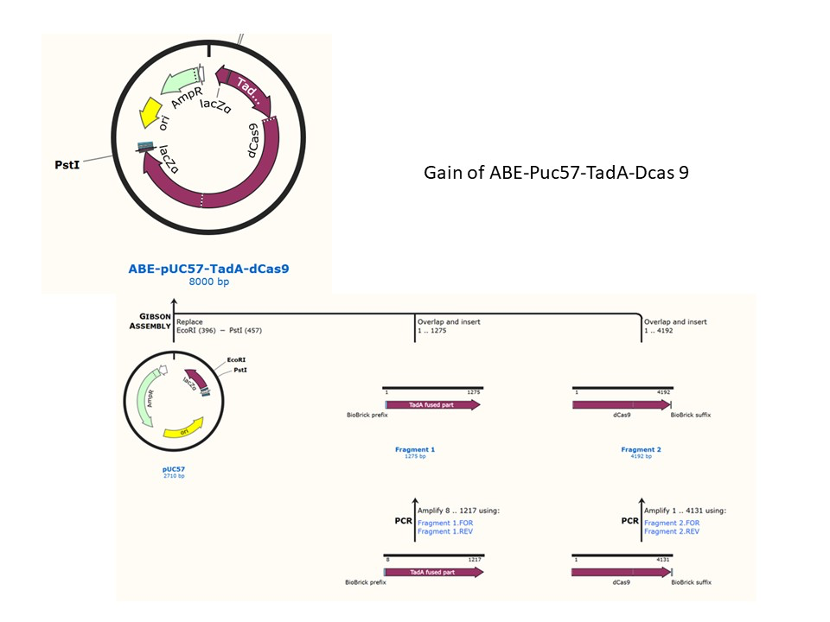
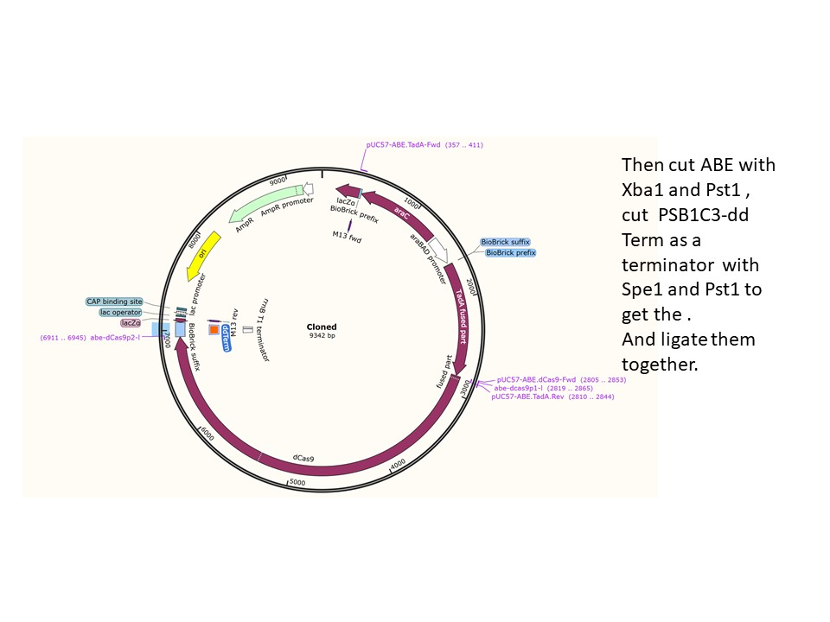
Citation
[1] Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, Bryant SH. " MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2014 Jan; 42(Database issue):D297-303
[2] Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017 Nov 23;551(7681):464-471. doi: 10.1038/nature24644. Epub 2017 Oct 25. Erratum in: Nature. 2018 May 2;: PMID: 29160308; PMCID: PMC5726555.
Note: due to Corona Pandemic and so limited experiment time, we were not able to characterize these parts.
Usage example2
designed by Peking 2022
Since discovered, Cas9 has been studied by many researchers to expand its application scope. Recently, the Evolvr system[1] which fused Cas9 with error-prone DNA Polymerase was developed and used for directed revolution in E. coli[2-4].As was reported in 2020, The group had proved the effectiveness of this system in yeast[5]. According to the research, Using a toxic arginine analogue to select S.cerevisiae that couldn't uptake this kind of chemicals due to a mutation in CAN1 gene (which is the target of EvolvR), researchers found that Evolvr was able to improve the targeted mutation rate in yeast by four order of magnitude elevation with weak genomic off-target effects.(Fig.1)

(
Fig.1 mutation rate of CAN1 for WT S.cerevisiae and Evolvr-carrying S.cerevisiae
(Tou CJ et al., 2020)

Fig.2 Mechanism of Evolvr system
(Tou CJ et al., 2020)
In recent years, EvolvR has been used by many researchers for directed evolution in E. coli [2-4], but its application in yeast was rarely reported. We gave it a try and added relevant information to the original Main Page as a usage extension for Cas9. In our work, we combined EvolvR with the co-culture system for directed evolution of yeast. Due to the limited time, our co-culture time was very short, but the sequencing results showed that the EPI sequence of few yeasts did change, which exactly interfered the function of target gene. (Fig.3,Fig.4). It takes more time to further verify and improve the function of EvolvR.

Fig.3 Sequencing results have shown a shift mutation in target sequence
< img src=" " width="50%" />
" width="50%" />
Fig.4 <p>Reference:
[1]. Halperin SO, Tou CJ, Wong EB, et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window[J]. Nature, 2018, 560(7717):248-52.
[2]. Garcia-Garcia JD, Joshi J, Patterson JA, et al. Potential for Applying Continuous Directed Evolution to Plant Enzymes: An Exploratory Study[J]. Life (Basel), 2020, 10(9).
[3]. Long M, Xu M, Qiao Z, et al. Directed Evolution of Ornithine Cyclodeaminase Using an EvolvR-Based Growth-Coupling Strategy for Efficient Biosynthesis of l-Proline[J]. ACS Synth Biol, 2020, 9(7):1855-63.
[4]. Rao GS, Jiang W, Mahfouz M. Synthetic directed evolution in plants: unlocking trait engineering and improvement[J]. Synth Biol (Oxf), 2021, 6(1):ysab025.
[5]. Tou CJ, Schaffer DV, Dueber JE. Targeted Diversification in the S. cerevisiae Genome with CRISPR-Guided DNA Polymerase I[J]. ACS Synth Biol, 2020, 9(7):1911-16.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 248
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_K1150000 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGACAAGAAG ... GGAGGCGACACCGGT ORF from nucleotide position -8 to 4107 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
References
[1] Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. (2012). The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 46, 311-39
[2] Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339(6121), 823-6
[3] Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 31(3), 233-9
[4] Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23
[5] Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5), 1173-83
