Difference between revisions of "Part:BBa K4162005"
(→Usage and Biology) |
|||
| Line 17: | Line 17: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | We construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between CDSs in a polycistron. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons in vivo. Self-interaction of the polycistron can be nullified and each cistron can initiate translation with comparable efficiency. Besides, we can precisely manage this co-expression system by adjusting the RBS strength of individual mono-cistrons. | |
===Characterization=== | ===Characterization=== | ||
Revision as of 11:21, 11 October 2022
Hammerhead ribozyme
Introduction
Hammerhead ribozyme was first found in the genome of viruses and viroids. It involved in the processing of RNA transcripts based on rolling-circle replication. The tandem copy of RNA sequence will be generated in the roll ring replication, and the self-cleaving activity of ribozyme can ensure the generation of RNA copy of unit length.[1]
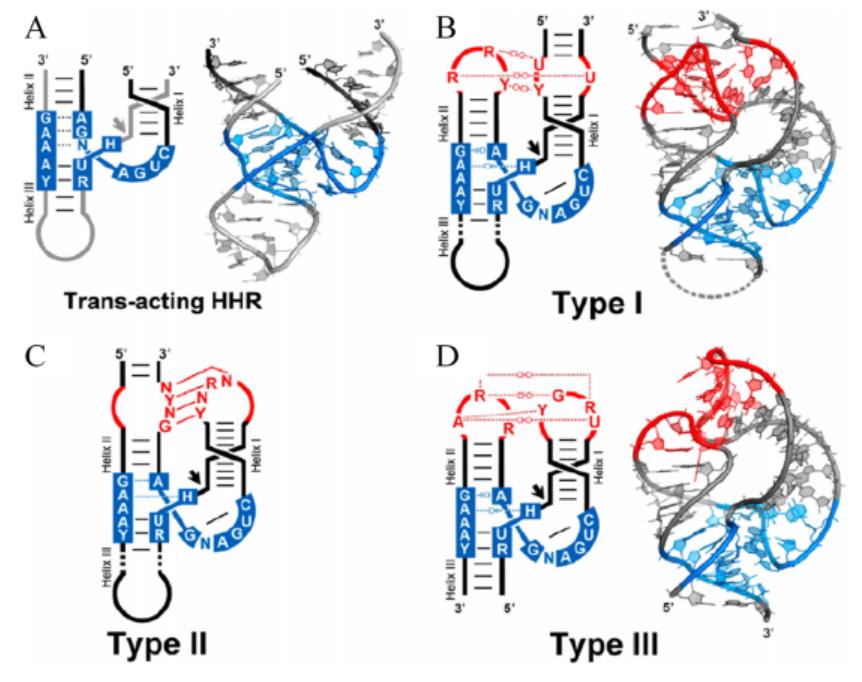
The secondary structure of hammerhead ribozyme resembles a hammer. According to different open helix tips, hammerhead ribozymes can be divided into three types: TypeⅠ, Type II and Type III. The catalytic center of ribozyme consists of 15 highly conserved bases surrounded by three helixs(HelixⅠ, Helix II and Helix III). The long-range interaction between HelixⅠ and Helix II can help stabilize the conformation of the catalytic center of the enzyme and improve the catalytic efficiency (Figure 1).[2] While working, ribozymes utilize a network of defined hydrogen bonds, ionic and hydrophobic interactions to generate catalytic pockets, which capitalize on steric constraints to generate in-line cleavage alignments and general acid-base chemistry to catalyze site-specific cleavage of the phosphodiester backbone.[3]
Contents
Usage and Biology
We construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between CDSs in a polycistron. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons in vivo. Self-interaction of the polycistron can be nullified and each cistron can initiate translation with comparable efficiency. Besides, we can precisely manage this co-expression system by adjusting the RBS strength of individual mono-cistrons.
Characterization
定性1标题
定性1文字
定性1小结
定性2标题
定性2文字
定性2小结
定性3标题
定性3文字
定性3小结
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Ferré-D'Amaré, A. R., & Scott, W. G. (2010). Small self-cleaving ribozymes. Cold Spring Harbor perspectives in biology, 2(10), a003574. https://doi.org/10.1101/cshperspect.a003574
- ↑ Jimenez, R. M., Polanco, J. A., & Lupták, A. (2015). Chemistry and Biology of Self-Cleaving Ribozymes. Trends in biochemical sciences, 40(11), 648–661. https://doi.org/10.1016/j.tibs.2015.09.001
- ↑ Ren, A., Micura, R., & Patel, D. J. (2017). Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. *Current opinion in chemical biology*, *41*, 71–83.