Difference between revisions of "Part:BBa K4162005"
| Line 40: | Line 40: | ||
====定性3标题 ==== | ====定性3标题 ==== | ||
| − | 定性3文字 | + | 定性3文字 |
[[File:T--Fudan--定性3图片不要用中文做文件名.png|400px|thumb|none|'''Figure 3. 图标题.''' 图注 引用的不要忘了写某某人某某年的出处]] | [[File:T--Fudan--定性3图片不要用中文做文件名.png|400px|thumb|none|'''Figure 3. 图标题.''' 图注 引用的不要忘了写某某人某某年的出处]] | ||
Revision as of 11:18, 11 October 2022
Hammerhead ribozyme
Introduction
Hammerhead ribozyme was first found in the genome of viruses and viroids. It involved in the processing of RNA transcripts based on rolling-circle replication. The tandem copy of RNA sequence will be generated in the roll ring replication, and the self-cleaving activity of ribozyme can ensure the generation of RNA copy of unit length.[1]
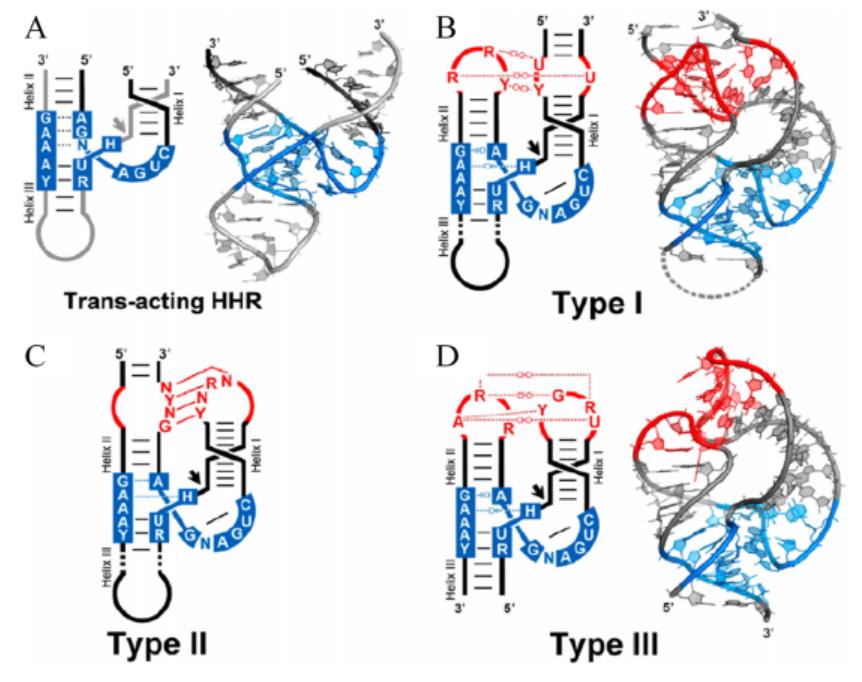
The secondary structure of hammerhead ribozyme resembles a hammer. According to different open helix tips, hammerhead ribozymes can be divided into three types: TypeⅠ, Type II and Type III. The catalytic center of ribozyme consists of 15 highly conserved bases surrounded by three helixs(HelixⅠ, Helix II and Helix III). The long-range interaction between HelixⅠ and Helix II can help stabilize the conformation of the catalytic center of the enzyme and improve the catalytic efficiency (Figure 1).[2] While working, ribozymes utilize a network of defined hydrogen bonds, ionic and hydrophobic interactions to generate catalytic pockets, which capitalize on steric constraints to generate in-line cleavage alignments and general acid-base chemistry to catalyze site-specific cleavage of the phosphodiester backbone.[3]
Contents
Usage and Biology
Hammerhead ribozyme can perform self-cleaving efficiently. While constructing polycistrons in E. coli, ribozyme sequence should be inserted between two coding sequences. After the transcription of CDS is completed, the ribozyme will conduct self-cleaving and separate the long piece of mRNA into segments corresponding to each CDS. Therefore, the translation process of target proteins will depend on their respective RBS strength.
Characterization
定性1标题
定性1文字
定性1小结
定性2标题
定性2文字
定性2小结
定性3标题
定性3文字
定性3小结
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Ferré-D'Amaré, A. R., & Scott, W. G. (2010). Small self-cleaving ribozymes. Cold Spring Harbor perspectives in biology, 2(10), a003574. https://doi.org/10.1101/cshperspect.a003574
- ↑ Jimenez, R. M., Polanco, J. A., & Lupták, A. (2015). Chemistry and Biology of Self-Cleaving Ribozymes. Trends in biochemical sciences, 40(11), 648–661. https://doi.org/10.1016/j.tibs.2015.09.001
- ↑ Ren, A., Micura, R., & Patel, D. J. (2017). Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. *Current opinion in chemical biology*, *41*, 71–83.