Difference between revisions of "Part:BBa K4380014"
| Line 14: | Line 14: | ||
<br><b>Properties</b> : Cell surface display protein system, which can be used as a immobilization unit on bacterial cellular membrane | <br><b>Properties</b> : Cell surface display protein system, which can be used as a immobilization unit on bacterial cellular membrane | ||
<br><b>Safety</b>: Biosafety level 1 laboratory | <br><b>Safety</b>: Biosafety level 1 laboratory | ||
| + | |||
| + | ====Usage==== | ||
| + | Bacterial display systems are routinely used for the improvement of antibodies, peptides, or enzymes. One frequently used bacterial cell surface display system is based on the EstA protein. Although there are different approaches on using this protein for cell surface display exposure, one of the most frequent uses involves a mutation at active residue S38A. This mutation eliminates enzymes esterase activity but the enzyme still able to be expressed on bacterial surfaces, therefore, can be used as an anchoring motif. The catalytically inactivated extracellular domain and the passenger domain are subsequently translocated to the cell surface [5][3] and ''E. coli'' cells can heterologously present approximately 36,000 copies of EstA on the cell surface[5]. | ||
| + | |||
| + | =====Directed evolution===== | ||
| + | ''E. coli'' cells present approximately 36,000 copies of EstA on the cell surface [5]. Therefore, high presence on bacterial surfaces (especially, when recombinant proteins can be expressed in high levels), is suitable for <b>Direct evolution approaches </b>. Direct Evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal[4]. This method has successfully transformed the way we view protein engineering. During ''in vivo'' evolution, each cell is transformed with a plasmid containing a different member of the variant library. Protein EstA allows to expression library variants on its surface, where its function can be tested. Therefore, enzymes that are presented on the cell surface or are secreted to the periplasm can successfully be engineered by directed evolution[6]. With this in mind, these systems are routinely used for the improvement of antibodies, peptides, enzymes, or nanobodies.[7][8][9] | ||
| + | |||
| + | ==Experimental characterisation== | ||
| + | |||
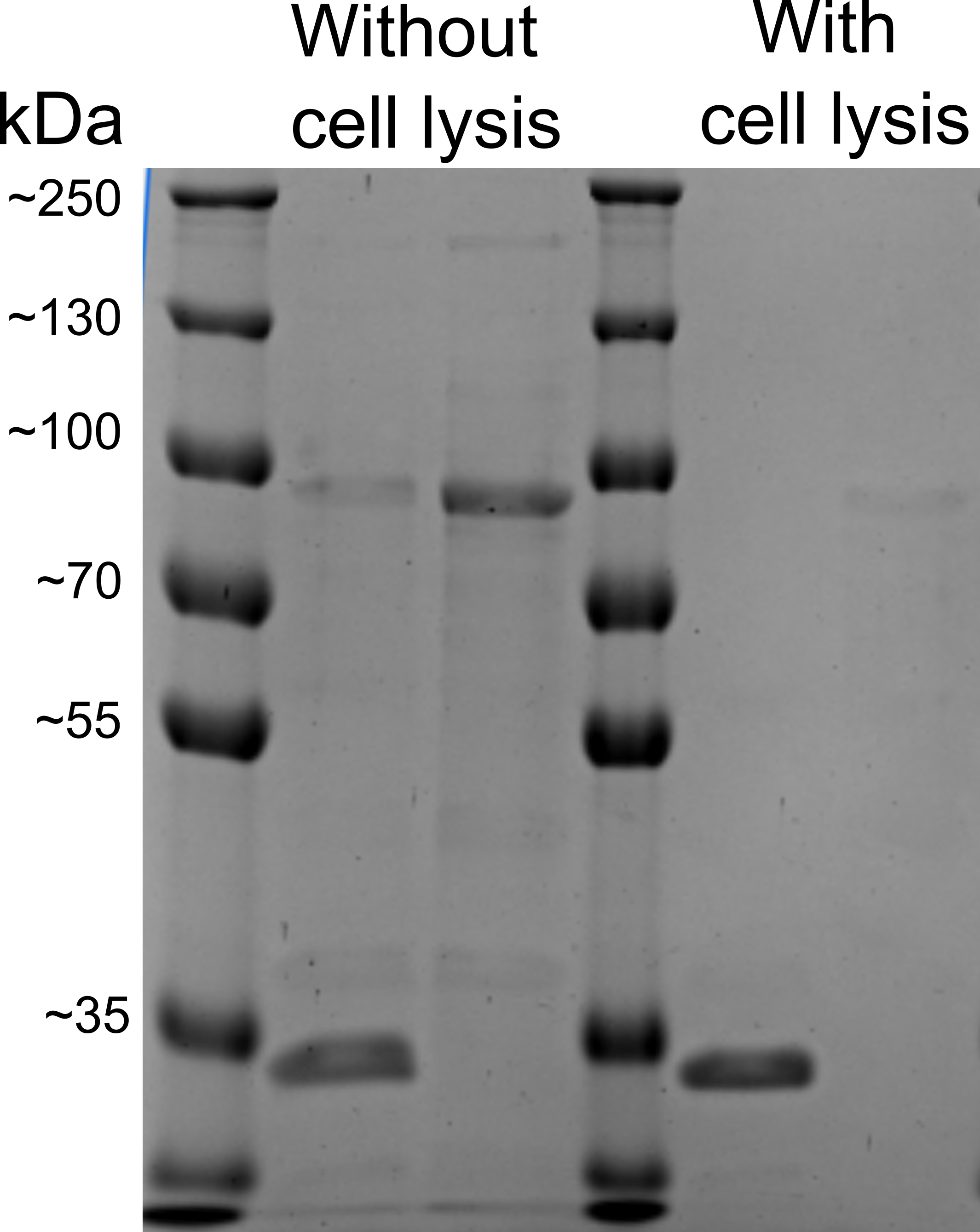
| + | During the development of the NanoFind detection system, we did encounter many problems, which needed creative and novel solutions. One of them was that our nanoplastic detection system might not be able to produce a signal, when nanoplastic concentrations are extremely low. To solve this, we might need a certain signal amplification system, if nanoplastics are present in the sample. [[File:pK.gif|200px|thumb|right|Figure 2: Proteinase K assay performed on cell surface display protein EstA with and without bacterial lysis]] | ||
Revision as of 03:49, 11 October 2022
EstA cell surface display system
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 444
Illegal BglII site found at 591
Illegal BamHI site found at 205 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 374
Illegal NgoMIV site found at 830
Illegal NgoMIV site found at 968
Illegal NgoMIV site found at 1508
Illegal NgoMIV site found at 1877 - 1000COMPATIBLE WITH RFC[1000]
Contents
Introduction
Vilnius-Lithuania Igem 2022 project NanoFind was working to create an easily accessible nanoplastic detection tool, using peptides, whose interaction with nanoplastic particles would lead to an easily interpretable response. The system itself focused on smaller protein molecules, peptides, which are modified to acquire the ability to connect to the surface of synthetic polymers – plastics. The detection system works when peptides and nanoplastic particles combine and form a sandwich complex - one nanoplastic particle is surrounded by two peptides, attached to their respective protein. The sandwich complex consisted of two main parts – one is a peptide bound to a fluorescent protein, and the other peptide is immobilized on a cellulose membrane by a cellulose binding domain.
A bacterial cell surface display system, based on the protein EstA was used as a novel way to expose the same peptides onto the bacterial membrane, allowing to amplify the signal of nanoplastic presence in a solution and finding novel peptides by using a peptide evolution protocol (PePevo). The team has generated several experimental characterizations, proving that the domain is successfully exposed on a bacterial surface, and created several new composite parts using this system.
Profile
Name: Bacterial cell surface display system, based on EstA protein from Pseudomonas aeruginosa
Origin: Synthetic, built from parts: BBa_K4380001 -> BBa_K4380006 -> BBa_K4380007 -> BBa_K4380005 -> BBa_K4380002 -> BBa_K2694001
Properties : Cell surface display protein system, which can be used as a immobilization unit on bacterial cellular membrane
Safety: Biosafety level 1 laboratory
Usage
Bacterial display systems are routinely used for the improvement of antibodies, peptides, or enzymes. One frequently used bacterial cell surface display system is based on the EstA protein. Although there are different approaches on using this protein for cell surface display exposure, one of the most frequent uses involves a mutation at active residue S38A. This mutation eliminates enzymes esterase activity but the enzyme still able to be expressed on bacterial surfaces, therefore, can be used as an anchoring motif. The catalytically inactivated extracellular domain and the passenger domain are subsequently translocated to the cell surface [5][3] and E. coli cells can heterologously present approximately 36,000 copies of EstA on the cell surface[5].
Directed evolution
E. coli cells present approximately 36,000 copies of EstA on the cell surface [5]. Therefore, high presence on bacterial surfaces (especially, when recombinant proteins can be expressed in high levels), is suitable for Direct evolution approaches . Direct Evolution (DE) is a method used in protein engineering that mimics the process of natural selection to steer proteins or nucleic acids toward a user-defined goal[4]. This method has successfully transformed the way we view protein engineering. During in vivo evolution, each cell is transformed with a plasmid containing a different member of the variant library. Protein EstA allows to expression library variants on its surface, where its function can be tested. Therefore, enzymes that are presented on the cell surface or are secreted to the periplasm can successfully be engineered by directed evolution[6]. With this in mind, these systems are routinely used for the improvement of antibodies, peptides, enzymes, or nanobodies.[7][8][9]