Difference between revisions of "Part:BBa K4140003"
Ahmed Mattar (Talk | contribs) (→Experimental Characterization) |
Ahmed Mattar (Talk | contribs) (→Experimental Characterization) |
||
| Line 24: | Line 24: | ||
Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase. | Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase. | ||
==Experimental Characterization== | ==Experimental Characterization== | ||
| − | [[File: | + | [[File:capture7.png|right|]] |
<br><br><br><br><br><br><br> | <br><br><br><br><br><br><br> | ||
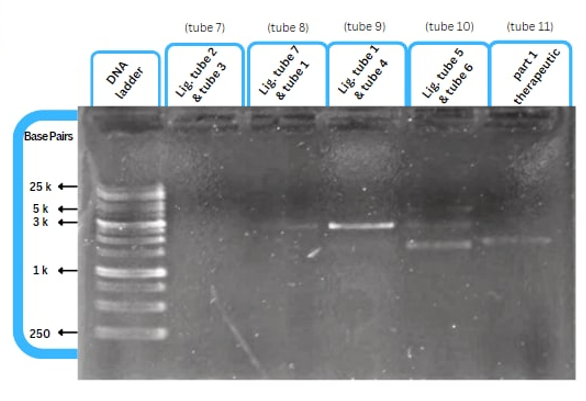
| − | This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane | + | This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 3. The run part (ordered from IDT) included ParoF promoter - P2A - L7Ae - KpSp - LacZ Alpha. |
<br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
Revision as of 16:03, 10 October 2022
ParoF promoter
Part Description
ParoF promoter is a constitutive promoter present in E. Coli which is sensitive to tyrosine and regulated by tyrR protein, it intitiates the synthesis of DAHP (3-deoxy-D-arabino-heptulosonate 7-phosphate synthase) and it is what makes it sensitive to phenylalanine as TyrR & Phenylalanine complex causes inhibition of the transcription of aroF
Usage
Due to its sensitivity to TyrR we use this promoter to regulate the expression of L7Ae under the effect of tyrR and phenylalanine the activity of paroF is optimum for L7Ae expression but in the presence of TyrR and tyrosine the expression of L7Ae is markedly reduced as shown in figure 1.
Characterization by mathematical modeling
We are using mathematical modeling to detect the increased level of phenylalanine (phe) in phenylketonuria patients in our diagnostic platform. It depends on a whole cell-based biosensor through a cascade of reactions to finally end by formation of β-galactosidase that turns the color into blue once bound to its substrate (X-gal) as mentioned in figure (1) and graph (1).
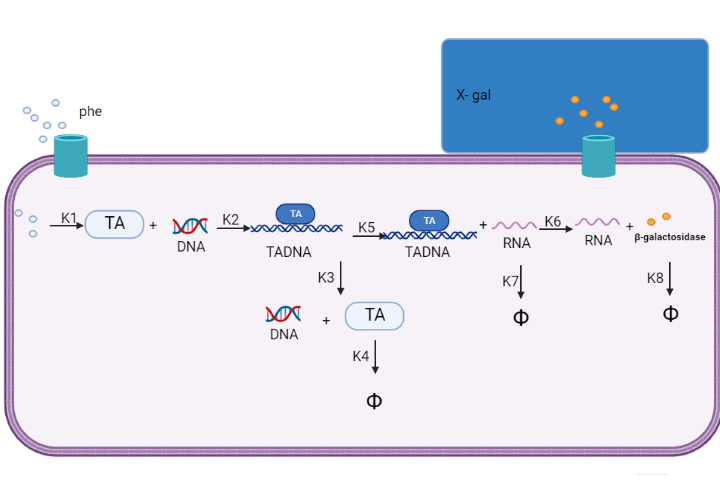
Figure (1) represents the cascade of reactions in whole cell-based biosensor model.
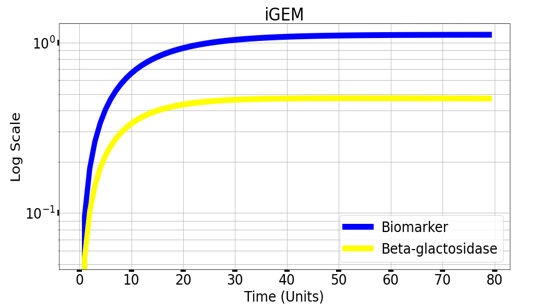
Graph(1) illustrates a direct relation between biomarker and beta-galactosidase ,so as the biomarker increases, the released amount of beta-galactosidase increases till it reaches constant value after about 30 time units. Therefore, the maximum amount of the biomarker releases the maximum amount of beta-galactosidase.
Experimental Characterization
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 3. The run part (ordered from IDT) included ParoF promoter - P2A - L7Ae - KpSp - LacZ Alpha.
References
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]