Difference between revisions of "Part:BBa K4140008"
Ahmed Mattar (Talk | contribs) |
Ahmed Mattar (Talk | contribs) (→Improvement by KP-SP tagging) |
||
| Line 19: | Line 19: | ||
=== Improvement by KP-SP tagging === | === Improvement by KP-SP tagging === | ||
| − | This year, we enhanced the LacZ alpha gene by incorporating a peptide signal that controls the secretion of B galactosidase extracellularly in an effort to improve the cleavage capacity of the X gal, heighten the intensity of the dark blue color emitted by the X gal product, and reduce the amount of time required to receive the results after performing the test on the chip | + | This year, we enhanced the LacZ alpha gene by incorporating a peptide signal that controls the secretion of B galactosidase extracellularly (KP-SP tagging) in an effort to improve the cleavage capacity of the X gal, heighten the intensity of the dark blue color emitted by the X gal product, and reduce the amount of time required to receive the results after performing the test on the chip. as shown in figure 2. |
| − | Improvement of LacZ alpha gene which encodes for the alpha domain of B-galactosidase that acts as a reporter protein in our | + | Improvement of LacZ alpha gene which encodes for the alpha domain of B-galactosidase that acts as a reporter protein in our diagnostic circuit : |
Lactose converting enzyme (B-gal) beta-galactosidase that is originated from bacteria And have been studied as a reporter protein for many years ,It's a polypeptide of 1029 amino acid, and its tetramerization carries out the functional form of the enzyme. | Lactose converting enzyme (B-gal) beta-galactosidase that is originated from bacteria And have been studied as a reporter protein for many years ,It's a polypeptide of 1029 amino acid, and its tetramerization carries out the functional form of the enzyme. | ||
| Line 27: | Line 27: | ||
The deletion of the tetramerization domain which is the N-terminal region spanning the first 50 residues will abolish its enzymatic activity, which could be fully restored by the alpha complementation of the intact N- terminal portion of the (Beta-gal) this alpha complementation is detected by using a beta-galactosidase colorless substrate as X gal to be cleaved into a dark blue product. | The deletion of the tetramerization domain which is the N-terminal region spanning the first 50 residues will abolish its enzymatic activity, which could be fully restored by the alpha complementation of the intact N- terminal portion of the (Beta-gal) this alpha complementation is detected by using a beta-galactosidase colorless substrate as X gal to be cleaved into a dark blue product. | ||
| − | This year we employed this approach to indicate the presence of phenylalanine within our whole cell biosensor on the test line with media containing X gal substrate of our platform to diagnose phenylketonuria | + | This year we employed this approach to indicate the presence of phenylalanine within our whole cell biosensor on the test line with media containing X gal substrate of our platform to diagnose phenylketonuria. |
| − | [[Image:Lacz_Improv.png|thumb|right|Figure( | + | [[Image:Lacz_Improv.png|thumb|right|Figure(2) Shows an SBOL demonstrating Lacz Improvement using Kp-Sp tagging ]] |
<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
Revision as of 09:56, 7 October 2022
lacZ alpha
Part Description
The LacZ-alpha fragment is encoded by this region and is derived from the pUC19 cloning vector. Because it is smaller than the LacZ-omega region, the LacZ-alpha fragment can be easily incorporated into a plasmid when the two non-functional LacZ gene fragments (alpha and omega) are co-expressed. X-gal (5-bromo-4-chloro-3-indoyl-d-galactopyranoside), a soluble colourless molecule that is a substrate of ß-galactosidase and generates a blue product when cleaved, is used to detect the alpha-complementation
Usage
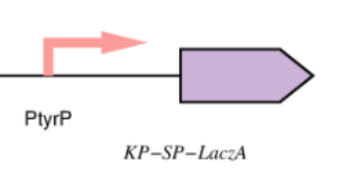
alpha domain of ß-galactosidase (our reporter protein) which is produced in response to high levels of phenylalanine and generates a blue product when X-gal is cleaved which is a soluble colorless molecule that is a substrate of ß-galactosidase and generates a blue product when cleaved, is used to detect the alpha-complementation. We used beta-galactosidase enzyme to turn the color into blue once increased level of phenylalanine is detected via exported extracellularly via tagging by kp-sp to bin with its substrate (X-gal) as shown in figure 1.
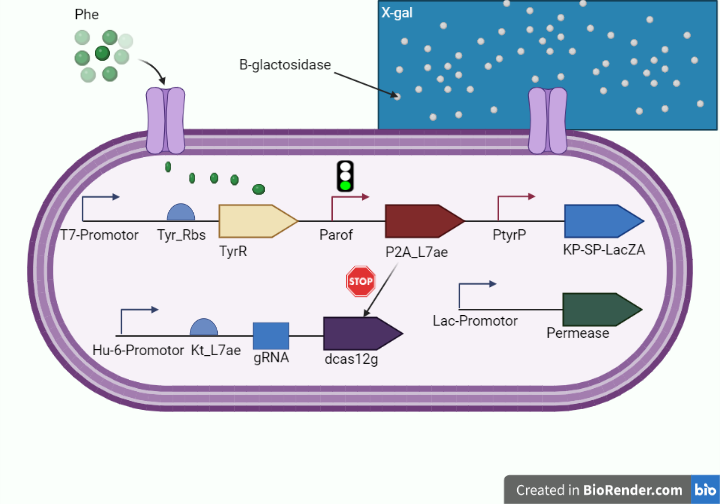
Figure 1. (shows the diagnostic whole cell-based biosensor.)
Improvements
We improved the part BBa_E0033 by :-
Improvement by KP-SP tagging
This year, we enhanced the LacZ alpha gene by incorporating a peptide signal that controls the secretion of B galactosidase extracellularly (KP-SP tagging) in an effort to improve the cleavage capacity of the X gal, heighten the intensity of the dark blue color emitted by the X gal product, and reduce the amount of time required to receive the results after performing the test on the chip. as shown in figure 2.
Improvement of LacZ alpha gene which encodes for the alpha domain of B-galactosidase that acts as a reporter protein in our diagnostic circuit :
Lactose converting enzyme (B-gal) beta-galactosidase that is originated from bacteria And have been studied as a reporter protein for many years ,It's a polypeptide of 1029 amino acid, and its tetramerization carries out the functional form of the enzyme.
The deletion of the tetramerization domain which is the N-terminal region spanning the first 50 residues will abolish its enzymatic activity, which could be fully restored by the alpha complementation of the intact N- terminal portion of the (Beta-gal) this alpha complementation is detected by using a beta-galactosidase colorless substrate as X gal to be cleaved into a dark blue product.
This year we employed this approach to indicate the presence of phenylalanine within our whole cell biosensor on the test line with media containing X gal substrate of our platform to diagnose phenylketonuria.
Improvement by Directed Evolution
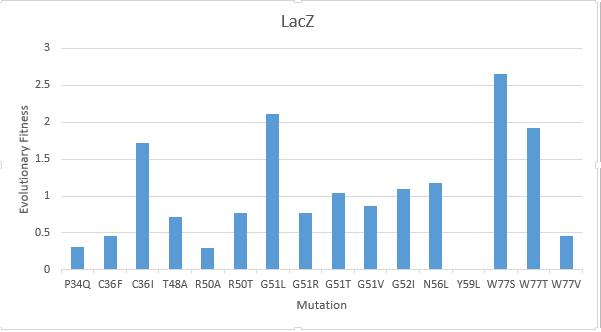
After creating a multiple sequence alignment of the protein sequence and predicting mutational landscapes, the effect of these mutations on the evolutionary fitness of the protein is tested. The prediction of the mutational landscape by saturation mutagenesis of the LacZ protein. The (W77S ) mutation, as depicted in the chart, had the greatest score when compared to other mutations. On the other hand, it's clear that the (Y59L) had the least evolutionary fitness for LacZ protein. As displayed in Figure(2)
Literature Characterization
According to the results, The ideal pH and temperature for Nf-LacZ are 40 °C and pH 6.5, respectively. The activity of the beta -galactosidase may also be impacted by metal ions. The metal ions K+, Mg2+, Ca2+, Zn2+, and Mn2+ were discovered to all be able to increase the enzymatic activity of Nf-LacZ.,and its Km was 05mmol/litre for Nf-LacZ, on the other hand the optimal temperature and PH for WspA are, respectively, 45 °C and pH 8.0, and Km value is close to to that of for WspA and this is showin in Nf-LacZ and WspA protein expression in vitro and analyzing its activity
References
1.Liu, W., Cui, L., Xu, H., Zhu, Z. & Gao, X. Flexibility-rigidity coordination of the dense exopolysaccharide matrix in terrestrial cyanobacteria acclimated to periodic desiccation. Appl. Environ. Microbiol. 83, e01619–17 (2017). 2.Wright, D. J. et al. UV irradiation and desiccation modulate the three-dimensional extracellular matrix of Nostoc commune (Cyanobacteria). J. Biol. Chem. 280, 40271–40281 (2005).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 93
Illegal XhoI site found at 144 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]