Difference between revisions of "Part:BBa K4247025"
| Line 1: | Line 1: | ||
===mCherry-SnoopCatcher=== | ===mCherry-SnoopCatcher=== | ||
| − | This part codes for the red fluorescent protein mCherry ( | + | This part codes for the red fluorescent protein mCherry (BBa_J06504) fused to SnoopCatcher (BBa_K4247009) via a linker. The SnoopCatcher enables the formation of a spontaneous isopeptide bond with any other protein containing the SnoopTag (BBa_K4247008). |
==Usage & Biology== | ==Usage & Biology== | ||
| − | Since the first successful cloning of the green fluorescent protein GFP of Aequorea victoria in 1992 (Prasher et al. 1992) fluorescent proteins became a widely used tool in many fields of research. Now an array of these proteins exists, with improved variants and a wide colour spectrum. | + | Since the first successful cloning of the green fluorescent protein (GFP) of Aequorea victoria in 1992 (Prasher et al. 1992) fluorescent proteins became a widely used tool in many fields of research. Now an array of these proteins exists, with improved variants and a wide colour spectrum. mCherry is a 26.7 kDa protein, and has a short maturation time (15 min) with an excitation max at 587 nm and emission max at 610 nm (www.fpbase.org). In 2006, its crystal structure was published (Shu and Remington 2006). |
| − | mCherry is a 26.7 kDa protein, and has a short maturation time (15 min) | + | |
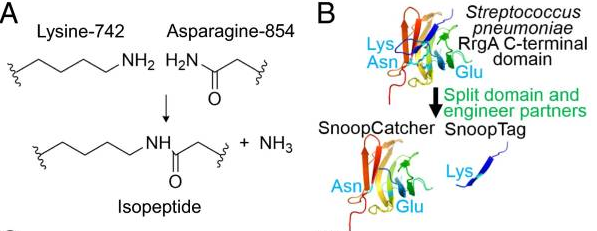
| − | + | A covalent peptide/protein pair was developed to enable spontaneous isopeptide bond formation between peptide tags. This system was developed based on Gram-positive surface protein, the pilus adhesin RrgA of S. pneumoniae. The D4 domain of this protein is stabilised by an isopeptide forming between a lysine (K742) and an asparagine (N854). This domain was split into a scaffold protein called SnoopCatcher and a 12-residue peptide termed SnoopTag, which can spontaneously form a covalent isopeptide bond upon mixing. The initiation, extension, and release steps use mild conditions, independent of redox state, and therefore should be applicable to a wide range of proteins according to the original paper. | |
| + | |||
| + | Here, we provide mCherry with an additional feature, the SnoopCatcher. After the fusion of SnoopCatcher, the mCherry protein has a MW of 40.9 kDa. The Tag-Catcher system is a protein conjugation tool that enables the formation of an irreversible isopeptide bond between these two components. | ||
[[File:Snoop 2.png]] | [[File:Snoop 2.png]] | ||
Revision as of 13:43, 30 September 2022
mCherry-SnoopCatcher
This part codes for the red fluorescent protein mCherry (BBa_J06504) fused to SnoopCatcher (BBa_K4247009) via a linker. The SnoopCatcher enables the formation of a spontaneous isopeptide bond with any other protein containing the SnoopTag (BBa_K4247008).
Usage & Biology
Since the first successful cloning of the green fluorescent protein (GFP) of Aequorea victoria in 1992 (Prasher et al. 1992) fluorescent proteins became a widely used tool in many fields of research. Now an array of these proteins exists, with improved variants and a wide colour spectrum. mCherry is a 26.7 kDa protein, and has a short maturation time (15 min) with an excitation max at 587 nm and emission max at 610 nm (www.fpbase.org). In 2006, its crystal structure was published (Shu and Remington 2006).
A covalent peptide/protein pair was developed to enable spontaneous isopeptide bond formation between peptide tags. This system was developed based on Gram-positive surface protein, the pilus adhesin RrgA of S. pneumoniae. The D4 domain of this protein is stabilised by an isopeptide forming between a lysine (K742) and an asparagine (N854). This domain was split into a scaffold protein called SnoopCatcher and a 12-residue peptide termed SnoopTag, which can spontaneously form a covalent isopeptide bond upon mixing. The initiation, extension, and release steps use mild conditions, independent of redox state, and therefore should be applicable to a wide range of proteins according to the original paper.
Here, we provide mCherry with an additional feature, the SnoopCatcher. After the fusion of SnoopCatcher, the mCherry protein has a MW of 40.9 kDa. The Tag-Catcher system is a protein conjugation tool that enables the formation of an irreversible isopeptide bond between these two components.