Difference between revisions of "Part:BBa K2872019"
(→Information contributed by City of London UK (2022)) |
|||
| (20 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2872019 short</partinfo> | <partinfo>BBa_K2872019 short</partinfo> | ||
| − | + | <partinfo>BBa_K2872019 SequenceAndFeatures</partinfo> | |
| + | ===Part Information=== | ||
| − | + | '''This part was synthesized using Cas12a DNA coding sequence from pY016 (pcDNA3.1-hLbCpf1) plasmid (Addgene plasmid # 69988) [1]''' | |
| − | + | <br> | |
| − | + | ===Biology and Usage=== | |
| − | < | + | |
| − | < | + | Cas12a is a programmable DNA endonuclease guided by a single guide RNA (gRNA) and can be used in Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems. RNA-guided DNA binding unleashes indiscriminate single-stranded DNA (ssDNA) cleavage activity by Cas12a that completely degrades ssDNA molecules [2] |
| + | |||
| + | *Targeting requires a gRNA complementary to the target site as well as a 5' TTTN protospacer adjacent motif (PAM) on the DNA strand opposite the target sequence. | ||
| + | *Cleavage occurs ~18 bases 3' of the PAM and leaves 5' overhanging ends. | ||
| + | *Cas12 a relies on a short (40-44 base) guide RNA to initiate DNA degradation. | ||
| + | *Cas12a is active from 16 to 48°C and maintains activity at lower temperatures than the Acidaminococcus orthologs, permitting editing in ectothermic organisms such as zebra fish and xenopus. | ||
| + | *High concentration liquid format can be used for microinjection, electroporation and lipofection [3] | ||
| + | |||
| + | There are 2 classes of effectors in the microbial adaptive immune system; class 1 effectors utilize multi-protein complexes, whereas class 2 effectors rely on single-component effector proteins such as the well-characterized Cas9. Cas12a (Cpf1) is a putative class 2 CRISPR effector which mediates robust DNA interference with features distinct from Cas9. Cpf1 is a single RNA-guided endonuclease lacking tracrRNA, and it utilizes a T-rich protospacer-adjacent motif. Moreover, Cpf1 cleaves DNA via a staggered DNA double-stranded break. We chose Lachnospiraceae bacterium ND2006 Cpf1 (LbCpf1) -along with Acidaminococcus- because it was found to have efficient genome-editing activity in human cells. [4] | ||
| + | |||
| + | |||
| + | ===Source=== | ||
| + | |||
| + | Lachnospiraceae bacterium ND2006 | ||
| + | |||
| + | ===References=== | ||
| + | [1] Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cell. 2015 Sep 23. pii: S0092-8674(15)01200-3. doi: 10.1016/j.cell.2015.09.038. 10.1016/j.cell.2015.09.038 PubMed 26422227 | ||
| + | <br> | ||
| + | [2] | ||
| + | Chen, J. S., Ma, E., Harrington, L. B., Costa, M. D., Tian, X., Palefsky, J. M., & Doudna, J. A. (2018, April 27). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Retrieved from http://science.sciencemag.org/content/360/6387/436 | ||
| + | <br> | ||
| + | [3] Biolabs, N. E. (n.d.). EnGen® Lba Cas12a (Cpf1). Retrieved from https://international.neb.com/products/m0653-engen-lba-cas12a-cpf1#Product Information_Notes | ||
| + | <br> | ||
| + | [4] Lassner, M. (2017). Faculty of 1000 evaluation for Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. F1000 - Post-publication Peer Review of the Biomedical Literature. doi:10.3410/f.725812264.793537694 | ||
| + | |||
| + | ===<h2 style="font-weight: bold;">iGEM Gifu 2021_Contribution</h2>=== | ||
| + | Group: iGEM Gifu 2021 | ||
| + | <br> | ||
| + | Author: Yuichiro IKAGAWA, Kyoka SATO <br> | ||
| + | <br> | ||
| + | ===Introduction=== | ||
| + | <html> | ||
| + | Cas12a recognizes and cleaves the target sequence of dsDNA depending on the target sequence and the T-rich PAM sequence. At this time, Cas12a exhibits collateral cleavage activity that indiscriminately cleaves the surrounding ssDNA. [1] CRISPR-AIOD assay, in which two different gRNAs are added, can improve the sensitivity. [2] The improvement of sensitivity by adding two different gRNAs has also been confirmed by an assay using Cas13a. [3] | ||
| + | |||
| + | </html> | ||
| + | <br><br> | ||
| + | ===Mechanism=== | ||
| + | <html> | ||
| + | Generally, in assays using a single gRNA, Cas is activated by the binding of a single Cas-gRNA complex to a single target DNA molecule. On the other hand, when two different gRNAs are introduced, two Cas-gRNA complexes bind to a single target DNA molecule, resulting in the activation of two Cas molecules. Since the number of Cas molecules activated per target molecule is doubled, sensitivity is expected to be improved [2], [3]. | ||
| + | |||
| + | </html> | ||
| + | <br> | ||
| + | |||
| + | ===Design=== | ||
| + | <html> | ||
| + | In the Cas12a-based CRISPR-AIOD assay, two target sequences are first set up on the DNA to be detected as targets. Different from the usual gRNA design, these two sequences are not restricted by the PAM sequence, but they must be close to each other. Next, two types of gRNAs are designed based on the set sequences, and primers that can bind upstream and downstream of the two target sites are designed. Finally, by adding the designed two types of gRNAs and primers together with recombinase polymerase amplification (RPA) reagent for target DNA, one-pot and highly sensitive detection becomes possible. As a result of introducing two types of gRNAs, it has been reported that a lower copy of DNA can be detected than when a single gRNA is added. [2]<br><br> | ||
| + | |||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center"> | ||
| + | <img src="https://2021.igem.org/wiki/images/5/56/T--Gifu--cas12a.png" style="width:50%"> | ||
| + | </div> | ||
| + | <br> | ||
| + | Figure 1. Mechanism of CRISPR Cas12a | ||
| + | <br><br> | ||
| + | |||
| + | When designing a gRNA with Cas13a, two target sequences on the RNA to be detected are set in the same way. In contrast to Cas12a, which does not require PAM to be taken into account when designing, in Cas13a, both gRNAs to be designed are restricted by protospacer-flanking site (PFS). Studies have shown that simultaneous introduction of the two designed gRNAs and Cas13a is possible even with low-copy RNAs, and the amount of signal detected per time has been improved. [3] | ||
| + | |||
| + | |||
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center"> | ||
| + | <img src="https://2021.igem.org/wiki/images/f/f7/T--Gifu--cas13a.png" style="width:50%"> | ||
| + | </div> | ||
| + | <br> | ||
| + | Figure 2. Mechanism of CRISPR Cas13a | ||
| + | <br> | ||
| + | </html> | ||
| + | <br> | ||
| + | |||
| + | ===Reference=== | ||
| + | [1]. Janice S. Chen, Enbo Ma, Lucas B. Harrington, Maria Da Costa, Xinran Tian, Joel M. Palefsky and Jennifer A. Doudna. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360, 436-439 (2018)<br> | ||
| + | [2]. Xiong Ding, Kun Yin, Ziyue Li, Rajesh V. Lalla, Enrique Ballesteros, Maroun M. Sfeir and Changchun Liu. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nature Communications, 11, 4711(2020)<br> | ||
| + | [3]. Parinaz Fozouni, Sungmin Son, María Díaz de León Derby, Gavin J Knott, Carley N Gray, Michael V D'Ambrosio, Chunyu Zhao, Neil A Switz, G. Renuka Kumar, Stephanie I Stephens et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323–333(2021)<br> | ||
| + | |||
| + | |||
| + | ===<h2 style="font-weight: bold;">Information contributed by City of London UK (2022)</h2>=== | ||
| + | Part information is collated here to help future users of the BioBrick registry. | ||
| + | |||
| + | Metadata: | ||
| + | *'''Group:''' City of London UK 2022 | ||
| + | *'''Author:''' Julian Chen, Elizabeth Noon | ||
| + | *'''Summary:''' Added information collated from existing scientific studies | ||
| + | |||
| + | ---- | ||
| + | |||
| + | Usage of the Cas12a enzyme may be integrated with lateral flow assay technology, alongside recombinase polymerase amplification (RPA). Using FAM-Biotin probes, a visible readout determining the presence of a gene of interest via a lateral flow assay can be achieved. The detection of RPA amplicons by gRNA/Cas complexes subsequently results in the activation of the Cas12a enzyme and commences the collateral trans-cleavage of FAM-Biotin (6-FAM/TTATTATT/Biotin) oligos, which are single-stranded DNA (ssDNA) probes, as Cas12a orthologs from the Lachnospiraceae family of bacteria (LbaCas12a) display non-specific collateral cleavage activity to ssDNA, post-detection of a gRNA-designated double-stranded DNA (dsDNA) target. <ref>Nguyen, P., Soenksen, L., Donghia, N., Angenent-Mari, N., de Puig, H., Huang, A., Lee, R., Slomovic, S., Galbersanini, T., Lansberry, G., Sallum, H., Zhao, E., Niemi, J. and Collins, J., 2021. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nature Biotechnology, 39(11), pp.1366-1374.</ref> | ||
| + | |||
| + | <html> | ||
| + | <img src="https://www.milenia-biotec.com/uploads/2020/04/Figure-3-1170x593.png" style="width:80%"> | ||
| + | </html> | ||
| + | <br> | ||
| + | Figure 3. Example of lateral flow assay technology combined with CRISPR Cas12a <ref>Milenia Biotec. 2022. Lateral Flow Readout for CRISPR/Cas-based detection strategies - Milenia Biotec. [online] Available at: <https://www.milenia-biotec.com/en/tips-lateral-flow-readouts-crispr-cas-strategies/> [Accessed 20 July 2022].</ref> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
| + | CRISPR Cas lateral flow assays use a reporter degradation strategy with the control ‘C’ line before the test ‘T’ line (a reversal compared to “standard” LFAs). When no CRISPR cas12a cleavage has occurred, gold nanoparticles conjugated to rabbit/goat anti-FAM antibodies bind to the uncleaved FAM/Biotin probes. These probes are immobilised at the control line as the biotin end binds to the Streptavidin tetramer present on the lateral flow strip, a natural protein-ligand interaction. When the FAM/Biotin probes have been cleaved the gold nanoparticles conjugated to the FAM end of the probe are free to flow to the test line where they are immobilised by anti-rabbit/goat antibodies. If both the T and C lines form, the result is considered positive but the more cleavage that has occurred the greater the strength of the T line and the weaker the strength of the C line. To ensure there is no signal at the T line when no cleavage has occurred, the correct amount of reporter must be used. Using too few reporter probes will leave some gold nanoparticles free to flow to the test line. However, using too many reporter probes relative to the number of binding sites can lead to the hook effect where excess probes bound to gold nanoparticles are no longer immobilised at the C line and so flow to the T line. <ref>2022. Confusion T and C Line. [ebook] Available at: <https://www.milenia-biotec.com/uploads/2019/07/Confusion-T-and-C-Line_final.pdf> [Accessed 20 July 2022].</ref> | ||
| + | <html> | ||
| + | <img src="https://www.milenia-biotec.com/uploads/2020/04/Figure-6_Reporter-induced-Hook-Effect-1140x611.png.webp" style="width:80%"> | ||
| + | </html> | ||
| + | <br> | ||
| + | Figure 4. Lateral flow assay using FAM-Biotin oligos as cleaved by Cas12 <ref>Milenia Biotec. 2022. Lateral Flow Readout for CRISPR/Cas-based detection strategies - Milenia Biotec. [online] Available at: <https://www.milenia-biotec.com/en/tips-lateral-flow-readouts-crispr-cas-strategies/> [Accessed 20 July 2022].</ref> | ||
| + | <br> | ||
| − | + | ===References=== | |
| − | === | + | |
| − | + | ||
| − | + | ||
Latest revision as of 09:25, 25 July 2022
Cas12a protein
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 1198
Illegal PstI site found at 1696
Illegal PstI site found at 3403 - 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 1198
Illegal PstI site found at 1696
Illegal PstI site found at 3403 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 938
Illegal BglII site found at 1271
Illegal BglII site found at 1841
Illegal BglII site found at 2108
Illegal BglII site found at 2729 - 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 1198
Illegal PstI site found at 1696
Illegal PstI site found at 3403 - 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 1198
Illegal PstI site found at 1696
Illegal PstI site found at 3403
Illegal NgoMIV site found at 1000
Illegal NgoMIV site found at 1999
Illegal NgoMIV site found at 2701 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1329
Illegal SapI.rc site found at 1389
Part Information
This part was synthesized using Cas12a DNA coding sequence from pY016 (pcDNA3.1-hLbCpf1) plasmid (Addgene plasmid # 69988) [1]
Biology and Usage
Cas12a is a programmable DNA endonuclease guided by a single guide RNA (gRNA) and can be used in Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems. RNA-guided DNA binding unleashes indiscriminate single-stranded DNA (ssDNA) cleavage activity by Cas12a that completely degrades ssDNA molecules [2]
- Targeting requires a gRNA complementary to the target site as well as a 5' TTTN protospacer adjacent motif (PAM) on the DNA strand opposite the target sequence.
- Cleavage occurs ~18 bases 3' of the PAM and leaves 5' overhanging ends.
- Cas12 a relies on a short (40-44 base) guide RNA to initiate DNA degradation.
- Cas12a is active from 16 to 48°C and maintains activity at lower temperatures than the Acidaminococcus orthologs, permitting editing in ectothermic organisms such as zebra fish and xenopus.
- High concentration liquid format can be used for microinjection, electroporation and lipofection [3]
There are 2 classes of effectors in the microbial adaptive immune system; class 1 effectors utilize multi-protein complexes, whereas class 2 effectors rely on single-component effector proteins such as the well-characterized Cas9. Cas12a (Cpf1) is a putative class 2 CRISPR effector which mediates robust DNA interference with features distinct from Cas9. Cpf1 is a single RNA-guided endonuclease lacking tracrRNA, and it utilizes a T-rich protospacer-adjacent motif. Moreover, Cpf1 cleaves DNA via a staggered DNA double-stranded break. We chose Lachnospiraceae bacterium ND2006 Cpf1 (LbCpf1) -along with Acidaminococcus- because it was found to have efficient genome-editing activity in human cells. [4]
Source
Lachnospiraceae bacterium ND2006
References
[1] Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cell. 2015 Sep 23. pii: S0092-8674(15)01200-3. doi: 10.1016/j.cell.2015.09.038. 10.1016/j.cell.2015.09.038 PubMed 26422227
[2]
Chen, J. S., Ma, E., Harrington, L. B., Costa, M. D., Tian, X., Palefsky, J. M., & Doudna, J. A. (2018, April 27). CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Retrieved from http://science.sciencemag.org/content/360/6387/436
[3] Biolabs, N. E. (n.d.). EnGen® Lba Cas12a (Cpf1). Retrieved from https://international.neb.com/products/m0653-engen-lba-cas12a-cpf1#Product Information_Notes
[4] Lassner, M. (2017). Faculty of 1000 evaluation for Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. F1000 - Post-publication Peer Review of the Biomedical Literature. doi:10.3410/f.725812264.793537694
iGEM Gifu 2021_Contribution
Group: iGEM Gifu 2021
Author: Yuichiro IKAGAWA, Kyoka SATO
Introduction
Cas12a recognizes and cleaves the target sequence of dsDNA depending on the target sequence and the T-rich PAM sequence. At this time, Cas12a exhibits collateral cleavage activity that indiscriminately cleaves the surrounding ssDNA. [1] CRISPR-AIOD assay, in which two different gRNAs are added, can improve the sensitivity. [2] The improvement of sensitivity by adding two different gRNAs has also been confirmed by an assay using Cas13a. [3]
Mechanism
Generally, in assays using a single gRNA, Cas is activated by the binding of a single Cas-gRNA complex to a single target DNA molecule. On the other hand, when two different gRNAs are introduced, two Cas-gRNA complexes bind to a single target DNA molecule, resulting in the activation of two Cas molecules. Since the number of Cas molecules activated per target molecule is doubled, sensitivity is expected to be improved [2], [3].
Design
In the Cas12a-based CRISPR-AIOD assay, two target sequences are first set up on the DNA to be detected as targets. Different from the usual gRNA design, these two sequences are not restricted by the PAM sequence, but they must be close to each other. Next, two types of gRNAs are designed based on the set sequences, and primers that can bind upstream and downstream of the two target sites are designed. Finally, by adding the designed two types of gRNAs and primers together with recombinase polymerase amplification (RPA) reagent for target DNA, one-pot and highly sensitive detection becomes possible. As a result of introducing two types of gRNAs, it has been reported that a lower copy of DNA can be detected than when a single gRNA is added. [2]

Figure 1. Mechanism of CRISPR Cas12a
When designing a gRNA with Cas13a, two target sequences on the RNA to be detected are set in the same way. In contrast to Cas12a, which does not require PAM to be taken into account when designing, in Cas13a, both gRNAs to be designed are restricted by protospacer-flanking site (PFS). Studies have shown that simultaneous introduction of the two designed gRNAs and Cas13a is possible even with low-copy RNAs, and the amount of signal detected per time has been improved. [3]

Figure 2. Mechanism of CRISPR Cas13a
Reference
[1]. Janice S. Chen, Enbo Ma, Lucas B. Harrington, Maria Da Costa, Xinran Tian, Joel M. Palefsky and Jennifer A. Doudna. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science, 360, 436-439 (2018)
[2]. Xiong Ding, Kun Yin, Ziyue Li, Rajesh V. Lalla, Enrique Ballesteros, Maroun M. Sfeir and Changchun Liu. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nature Communications, 11, 4711(2020)
[3]. Parinaz Fozouni, Sungmin Son, María Díaz de León Derby, Gavin J Knott, Carley N Gray, Michael V D'Ambrosio, Chunyu Zhao, Neil A Switz, G. Renuka Kumar, Stephanie I Stephens et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323–333(2021)
Information contributed by City of London UK (2022)
Part information is collated here to help future users of the BioBrick registry.
Metadata:
- Group: City of London UK 2022
- Author: Julian Chen, Elizabeth Noon
- Summary: Added information collated from existing scientific studies
Usage of the Cas12a enzyme may be integrated with lateral flow assay technology, alongside recombinase polymerase amplification (RPA). Using FAM-Biotin probes, a visible readout determining the presence of a gene of interest via a lateral flow assay can be achieved. The detection of RPA amplicons by gRNA/Cas complexes subsequently results in the activation of the Cas12a enzyme and commences the collateral trans-cleavage of FAM-Biotin (6-FAM/TTATTATT/Biotin) oligos, which are single-stranded DNA (ssDNA) probes, as Cas12a orthologs from the Lachnospiraceae family of bacteria (LbaCas12a) display non-specific collateral cleavage activity to ssDNA, post-detection of a gRNA-designated double-stranded DNA (dsDNA) target. [1]
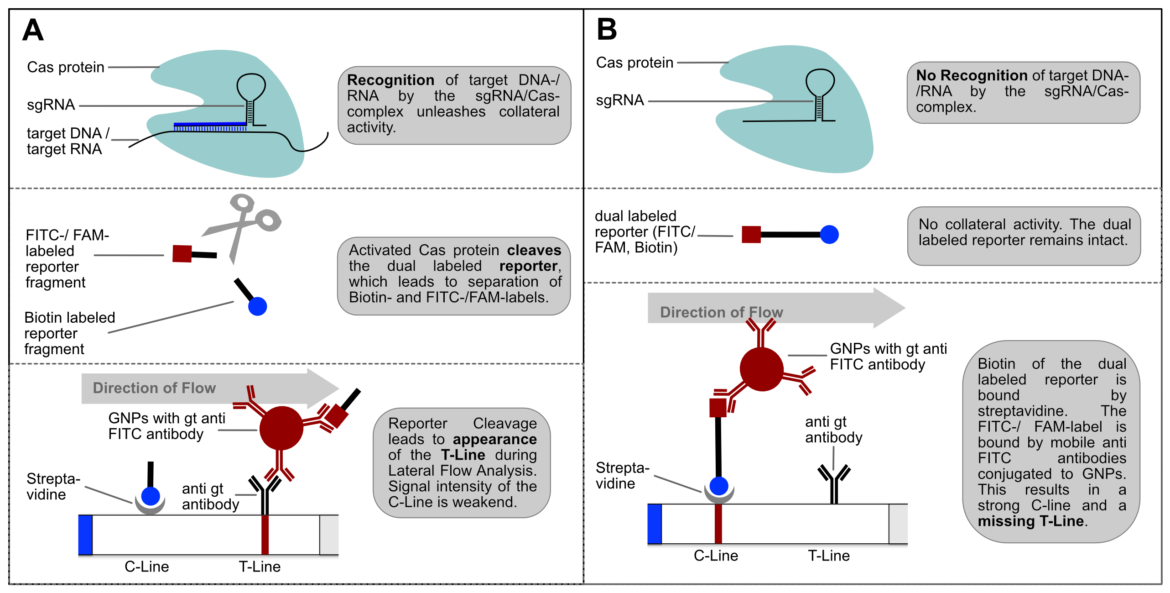
Figure 3. Example of lateral flow assay technology combined with CRISPR Cas12a [2]
CRISPR Cas lateral flow assays use a reporter degradation strategy with the control ‘C’ line before the test ‘T’ line (a reversal compared to “standard” LFAs). When no CRISPR cas12a cleavage has occurred, gold nanoparticles conjugated to rabbit/goat anti-FAM antibodies bind to the uncleaved FAM/Biotin probes. These probes are immobilised at the control line as the biotin end binds to the Streptavidin tetramer present on the lateral flow strip, a natural protein-ligand interaction. When the FAM/Biotin probes have been cleaved the gold nanoparticles conjugated to the FAM end of the probe are free to flow to the test line where they are immobilised by anti-rabbit/goat antibodies. If both the T and C lines form, the result is considered positive but the more cleavage that has occurred the greater the strength of the T line and the weaker the strength of the C line. To ensure there is no signal at the T line when no cleavage has occurred, the correct amount of reporter must be used. Using too few reporter probes will leave some gold nanoparticles free to flow to the test line. However, using too many reporter probes relative to the number of binding sites can lead to the hook effect where excess probes bound to gold nanoparticles are no longer immobilised at the C line and so flow to the T line. [3]

Figure 4. Lateral flow assay using FAM-Biotin oligos as cleaved by Cas12 [4]
References
- ↑ Nguyen, P., Soenksen, L., Donghia, N., Angenent-Mari, N., de Puig, H., Huang, A., Lee, R., Slomovic, S., Galbersanini, T., Lansberry, G., Sallum, H., Zhao, E., Niemi, J. and Collins, J., 2021. Wearable materials with embedded synthetic biology sensors for biomolecule detection. Nature Biotechnology, 39(11), pp.1366-1374.
- ↑ Milenia Biotec. 2022. Lateral Flow Readout for CRISPR/Cas-based detection strategies - Milenia Biotec. [online] Available at: <https://www.milenia-biotec.com/en/tips-lateral-flow-readouts-crispr-cas-strategies/> [Accessed 20 July 2022].
- ↑ 2022. Confusion T and C Line. [ebook] Available at: <https://www.milenia-biotec.com/uploads/2019/07/Confusion-T-and-C-Line_final.pdf> [Accessed 20 July 2022].
- ↑ Milenia Biotec. 2022. Lateral Flow Readout for CRISPR/Cas-based detection strategies - Milenia Biotec. [online] Available at: <https://www.milenia-biotec.com/en/tips-lateral-flow-readouts-crispr-cas-strategies/> [Accessed 20 July 2022].
