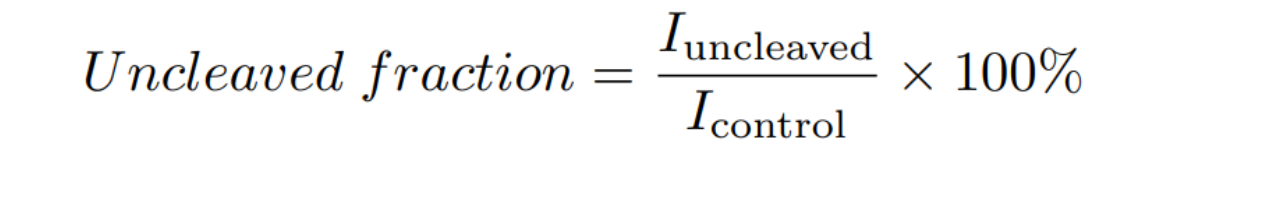Difference between revisions of "Part:BBa K3806014"
| (37 intermediate revisions by 3 users not shown) | |||
| Line 7: | Line 7: | ||
==Usage and Biology== | ==Usage and Biology== | ||
| − | Sensing small molecules is the foundation of many applications, ranging from disease diagnosis, prognosis, or treatment to detecting small pollutants in the environment. Synthetic genetic switches are a promising tool to detect and quantify small molecules. Conventional synthetic biology-based biosensors | + | Sensing small molecules is the foundation of many applications, ranging from disease diagnosis, prognosis, or treatment to detecting small pollutants in the environment. Synthetic genetic switches are a promising tool to detect and quantify small molecules. Conventional synthetic biology-based biosensors are based on transcription factors. However, transcription factors are not easily reprogrammable with respect to ligand selectivity. Antibody-based biosensors can be easily developed into new sensing capabilities. Unfortunately, these types of sensors are not suitable to detect low molecular weight compounds. RNA-based biosensors are an interesting alternative since they present a remarkable flexibility to be engineered into sensing a wide range of analytes with high sensitivity and selectivity. In particular, aptazymes, ligand regulated self-cleaving ribozymes, are of special interest as cleavage of the aptazyme can be coupled to regulated gene expression <i>in vivo</i> or <i>in vitro</i>. Moreover, newly developed methods such as DRIVER (<i>de novo</i> rapid <i>in vitro</i> evolution of RNA biosensors) enable rapid, automated, and multiplexed engineering of aptazymes sequences to diverse ligands [1]. |
| − | The TU Delft 2021 team provides the iGEM community with a novel genetic switch construct (BBa_K3806014) in which an aptazyme sequence is fused to the <i>lacZ</i> reporter gene | + | The TU Delft 2021 team provides the iGEM community with a novel genetic switch construct (<HTML><a href="https://parts.igem.org/Part:BBa_K3806014" target="_blank"><b>BBa_K3806014</b></a></HTML>) in which an aptazyme sequence is fused to the <i>lacZ</i> reporter gene to covert ligand concentration to a colorimetric read-out. The construct is designed to be modular and serve as a template to engineer other ligand-specific genetic circuits by swapping the aptazyme domain. It was demonstrated that the designed genetic construct can be expressed in a cell-free system by using a known theophylline binding aptazyme [1]. |
<HTML><h3>Aptazyme-regulated gene expression mechanism</h3></html> | <HTML><h3>Aptazyme-regulated gene expression mechanism</h3></html> | ||
| − | The aptazyme-regulated expression of <i>lacZ</i> depends on the accessibility of the ribosomal binding site (RBS) for translation initiation | + | The aptazyme-regulated expression of <i>lacZ</i> depends on the accessibility of the ribosomal binding site (RBS) for translation initiation as inspired by the study of Klauser & Hartig [2]. After transcription of the DNA template comprising the aptazyme and fused <i>lacZ</i> reporter gene, the RBS is hidden in the stem of the aptazyme. When the aptazyme is stabilized upon binding of the ligand, the RBS remains entirely sequestered by its antisense strand, and translation is mostly prohibited (Fig. 1, left). Cleavage of the aptazyme in the absence of the ligand liberates the RBS. As a result, the ribosome can bind to the RBS and translate the downstream reporter gene to the β-galactosidase protein (Fig. 1, right). Subsequently, the β-galactosidase enzyme can catalyze the colorimetric conversion of enzymatic substrates such as chlorophenol red-b-D-galactopyranoside (CPRG) and X-gal. |
| − | + | [[File:T--TUDelft--Mech_parts_cRBS.jpg|800px|center|]] | |
| + | <html><b><i>Fig. 1 Aptazyme-regulated gene expression mechanism.</i></b> Binding of the ligand renders a catalytically inactive aptazyme, the RBS remains entirely sequestered by its antisense strand, repressing translation (left). In the absence of the ligand, self-cleavage of the aptazyme frees the RBS, resulting in the binding of the small ribosomal subunit and initiation of translation (right). </html> | ||
| − | + | ==Experimental results== | |
| − | + | ||
| − | + | ||
| − | == | + | |
<HTML><h3>Cleavage characterization of the aptazyme-regulated genetic construct in T7 transcription reaction</h3> | <HTML><h3>Cleavage characterization of the aptazyme-regulated genetic construct in T7 transcription reaction</h3> | ||
</html> | </html> | ||
| − | + | To assess the ligand-dependent cleavage activity of the theophylline aptazyme in <HTML><a href="https://parts.igem.org/Part:BBa_K3806014" target="_blank"><b>BBa_K3806014</b></a></HTML>, the aptazyme-fused <i>lacZ</i> transcript product was analysed by RT-PCR. First the genetic construct was transcribed in a T7 transcription reaction for 1 hour (Fig. 2, step 1) followed by a reverse transcription (RT) reaction to convert the transcribed mRNA to cDNA (step 2). These cDNA products were then amplified using two primer sets. One that flanks the cleavage site (uncleaved region) and one that flanks a region downstream the cleavage site (control region). In the former case, it should be noted that only the uncleaved mRNA fraction will be PCR amplified (step 3). As a final step, the PCR product was visualized on an agarose gel (step 4). A lower band intensity is expected with decreasing concentration of theophylline, meaning that aptazyme self-cleaved and the forward primer flanking the cleavage site was unable to bind to the 5’ of the cDNA to produce a PCR product. The region downstream the cleavage point was used as control for transcription, RT, PCR and loading in the agarose gel, as band intensity should be similar between samples regardless of theophylline addition. | |
| + | [[File:T--TUDelft--RTqPCR_cRBS.jpg|900px|center|]] | ||
| − | + | <HTML><b><i>Fig. 2 Scheme of the 4 steps of the cleavage characterization procedure (left). Agarose gel analysis of the theophylline-dependent cleavage activity of the aptazyme after T7 transcription (right). </b> (A) PCR product after amplification of the uncleaved region (the uncleaved fraction is displayed in the bottom of the gel). (B) PCR product after amplification of the control region. Lanes: (Ladder) dsDNA ladder. (1) NC: negative control without DNA template. (2) 0 mM theophylline (3) 1 mM theophylline. (4) 5 mM theophylline. </i></html> | |
| − | [[File:T--TUDelft-- | + | ImageJ was used to quantify the intensity (I) of the gel bands, and the uncleaved fraction was determined by: |
| − | + | [[File:T--TUDelft--unCleaved f.png|500px|center|]] | |
| − | The | + | The computed uncleaved fractions were 36%, 52%, and 60% for 0, 1 and 5 mM theophylline, respectively (Fig. 2A). From this it can be concluded that the aptazyme presents theophylline-dependent cleavage activity in the T7 transcription buffer, and that the fusion of the <i>lacZ</i> gene to the aptazyme sequence does not hinder aptazyme self-cleavage. |
| + | |||
| + | <HTML><h3>Expression of genetic construct in absence and presence of ligand</h3></html> | ||
| + | For a colorimetric characterization of <HTML><a href="https://parts.igem.org/Part:BBa_K3806014" target="_blank"><b>BBa_K3806014</b></a></HTML>, this part was expressed in PURExpress using CPRG as a substrate. As expected, a decrease in expression can be seen when theophylline is added to the reaction (Fig. 3). | ||
| + | |||
| + | [[File:T--TUDelft--PURExpressCPRG.png|300px|center|]] | ||
| + | |||
| + | <HTML><b><i>Fig. 3 Image of PURExpress reactions using CPRG as substrate following 1 hour incubation.</i> </b>Samples: (1) without DNA template and 0 mM theophylline, yellow; (2) without DNA template and 6 mM theophylline, yellow; (3) with DNA template and 0 mM theophylline, red; and (4) with DNA template and 6 mM theophylline, orange. The DNA template was added at a final concentration of 5 nM. CPRG was used as a substrate to a final concentration of 0.6 mg/ml. </html> | ||
| + | |||
| + | Additionally, the product formation was determined by measuring the CPR absorbance at 575 nm (Fig. 3). Decreased absorbance is again observed at 5 mM concentration of theophylline compared to 0 mM theophylline. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
[[File:T--TUDelft--kinetics.png|500px|center|]] | [[File:T--TUDelft--kinetics.png|500px|center|]] | ||
| − | <HTML><b><i>Fig | + | <HTML><b><i>Fig 4 Expression of BBa_K3806014 in PURExpress with and wihout the addition of theophylline.</i></b> CPR production was quantified as a measure of absorbance at 575 nm.</html> |
| − | |||
<HTML><h3>Ligand-regulated expression on paper</h3></html> | <HTML><h3>Ligand-regulated expression on paper</h3></html> | ||
| − | Next, the ligand-regulated expression system was tested on a paper support. Cell-free system reactions were | + | Next, the ligand-regulated expression system was tested on a paper support. Cell-free system reactions were assembled and applied into paper discs, and incubated at 37 °C for 24 hours (Fig. 4). After 24 hours of expression, an increased color change is visible for the sample without theophylline, compared to the 5 mM theophylline (Fig. 4, 24 h). This is in correspondence with the results observed in the previous experiment in liquid. |
| − | [[File:T--TUDelft--paper.png| | + | [[File:T--TUDelft--paper.png|300px|center|]] |
| − | <HTML><b><i>Fig | + | <HTML><b><i>Fig 5 Paper-based reactions using PURExpress after reaction assembly (0 h), and after 24 hours of expression at 37 °C (24 h).</i></b> The part was used at a concentration of 3 nM. From left to right: (1) without DNA template and 0 mM theophylline, (2) with DNA template and 0 mM theophylline, and (3) with DNA template and 5 mM theophylline. CPRG was used as a substrate to a final concentration of 0.6 mg/ml.</html> |
| − | === | + | Although, <html><a href="https://2021.igem.org/Team:TUDelft/Results">alternative experiments</a></html> using a control part without an aptazyme sequence showed inhibitory effects of theophylline on the PURE cell-free system, the expression profiles and the cleavage characterization of <HTML><a href="https://parts.igem.org/Part:BBa_K3806014" target="_blank"><b>BBa_K3806014</b></a><HTML>, support that the <i>lacZ</i> expression from this part is ligand-responsive. For an improved version of this part, check <HTML><a href="https://parts.igem.org/Part:BBa_K3806016" target="_blank"><b>BBa_K3806016</b></a></HTML>. |
| + | |||
| + | |||
| + | ==References== | ||
<html> | <html> | ||
<style> | <style> | ||
| Line 71: | Line 76: | ||
</ol> | </ol> | ||
| − | <html> | + | </html> |
| + | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K3806008 parameters</partinfo> |
<!-- --> | <!-- --> | ||
Latest revision as of 22:09, 21 October 2021
Theophylline-binding aptazyme regulating lacZ expression (cRBS). With T7 promoter.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3204
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2321
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Sensing small molecules is the foundation of many applications, ranging from disease diagnosis, prognosis, or treatment to detecting small pollutants in the environment. Synthetic genetic switches are a promising tool to detect and quantify small molecules. Conventional synthetic biology-based biosensors are based on transcription factors. However, transcription factors are not easily reprogrammable with respect to ligand selectivity. Antibody-based biosensors can be easily developed into new sensing capabilities. Unfortunately, these types of sensors are not suitable to detect low molecular weight compounds. RNA-based biosensors are an interesting alternative since they present a remarkable flexibility to be engineered into sensing a wide range of analytes with high sensitivity and selectivity. In particular, aptazymes, ligand regulated self-cleaving ribozymes, are of special interest as cleavage of the aptazyme can be coupled to regulated gene expression in vivo or in vitro. Moreover, newly developed methods such as DRIVER (de novo rapid in vitro evolution of RNA biosensors) enable rapid, automated, and multiplexed engineering of aptazymes sequences to diverse ligands [1].
The TU Delft 2021 team provides the iGEM community with a novel genetic switch construct (BBa_K3806014) in which an aptazyme sequence is fused to the lacZ reporter gene to covert ligand concentration to a colorimetric read-out. The construct is designed to be modular and serve as a template to engineer other ligand-specific genetic circuits by swapping the aptazyme domain. It was demonstrated that the designed genetic construct can be expressed in a cell-free system by using a known theophylline binding aptazyme [1].
Aptazyme-regulated gene expression mechanism
The aptazyme-regulated expression of lacZ depends on the accessibility of the ribosomal binding site (RBS) for translation initiation as inspired by the study of Klauser & Hartig [2]. After transcription of the DNA template comprising the aptazyme and fused lacZ reporter gene, the RBS is hidden in the stem of the aptazyme. When the aptazyme is stabilized upon binding of the ligand, the RBS remains entirely sequestered by its antisense strand, and translation is mostly prohibited (Fig. 1, left). Cleavage of the aptazyme in the absence of the ligand liberates the RBS. As a result, the ribosome can bind to the RBS and translate the downstream reporter gene to the β-galactosidase protein (Fig. 1, right). Subsequently, the β-galactosidase enzyme can catalyze the colorimetric conversion of enzymatic substrates such as chlorophenol red-b-D-galactopyranoside (CPRG) and X-gal.Fig. 1 Aptazyme-regulated gene expression mechanism. Binding of the ligand renders a catalytically inactive aptazyme, the RBS remains entirely sequestered by its antisense strand, repressing translation (left). In the absence of the ligand, self-cleavage of the aptazyme frees the RBS, resulting in the binding of the small ribosomal subunit and initiation of translation (right).
Experimental results
Cleavage characterization of the aptazyme-regulated genetic construct in T7 transcription reaction
To assess the ligand-dependent cleavage activity of the theophylline aptazyme in BBa_K3806014, the aptazyme-fused lacZ transcript product was analysed by RT-PCR. First the genetic construct was transcribed in a T7 transcription reaction for 1 hour (Fig. 2, step 1) followed by a reverse transcription (RT) reaction to convert the transcribed mRNA to cDNA (step 2). These cDNA products were then amplified using two primer sets. One that flanks the cleavage site (uncleaved region) and one that flanks a region downstream the cleavage site (control region). In the former case, it should be noted that only the uncleaved mRNA fraction will be PCR amplified (step 3). As a final step, the PCR product was visualized on an agarose gel (step 4). A lower band intensity is expected with decreasing concentration of theophylline, meaning that aptazyme self-cleaved and the forward primer flanking the cleavage site was unable to bind to the 5’ of the cDNA to produce a PCR product. The region downstream the cleavage point was used as control for transcription, RT, PCR and loading in the agarose gel, as band intensity should be similar between samples regardless of theophylline addition.Fig. 2 Scheme of the 4 steps of the cleavage characterization procedure (left). Agarose gel analysis of the theophylline-dependent cleavage activity of the aptazyme after T7 transcription (right). (A) PCR product after amplification of the uncleaved region (the uncleaved fraction is displayed in the bottom of the gel). (B) PCR product after amplification of the control region. Lanes: (Ladder) dsDNA ladder. (1) NC: negative control without DNA template. (2) 0 mM theophylline (3) 1 mM theophylline. (4) 5 mM theophylline.
ImageJ was used to quantify the intensity (I) of the gel bands, and the uncleaved fraction was determined by:
The computed uncleaved fractions were 36%, 52%, and 60% for 0, 1 and 5 mM theophylline, respectively (Fig. 2A). From this it can be concluded that the aptazyme presents theophylline-dependent cleavage activity in the T7 transcription buffer, and that the fusion of the lacZ gene to the aptazyme sequence does not hinder aptazyme self-cleavage.
Expression of genetic construct in absence and presence of ligand
For a colorimetric characterization of BBa_K3806014, this part was expressed in PURExpress using CPRG as a substrate. As expected, a decrease in expression can be seen when theophylline is added to the reaction (Fig. 3).Fig. 3 Image of PURExpress reactions using CPRG as substrate following 1 hour incubation. Samples: (1) without DNA template and 0 mM theophylline, yellow; (2) without DNA template and 6 mM theophylline, yellow; (3) with DNA template and 0 mM theophylline, red; and (4) with DNA template and 6 mM theophylline, orange. The DNA template was added at a final concentration of 5 nM. CPRG was used as a substrate to a final concentration of 0.6 mg/ml.
Additionally, the product formation was determined by measuring the CPR absorbance at 575 nm (Fig. 3). Decreased absorbance is again observed at 5 mM concentration of theophylline compared to 0 mM theophylline.
Fig 4 Expression of BBa_K3806014 in PURExpress with and wihout the addition of theophylline. CPR production was quantified as a measure of absorbance at 575 nm.
Ligand-regulated expression on paper
Next, the ligand-regulated expression system was tested on a paper support. Cell-free system reactions were assembled and applied into paper discs, and incubated at 37 °C for 24 hours (Fig. 4). After 24 hours of expression, an increased color change is visible for the sample without theophylline, compared to the 5 mM theophylline (Fig. 4, 24 h). This is in correspondence with the results observed in the previous experiment in liquid.
Fig 5 Paper-based reactions using PURExpress after reaction assembly (0 h), and after 24 hours of expression at 37 °C (24 h). The part was used at a concentration of 3 nM. From left to right: (1) without DNA template and 0 mM theophylline, (2) with DNA template and 0 mM theophylline, and (3) with DNA template and 5 mM theophylline. CPRG was used as a substrate to a final concentration of 0.6 mg/ml.
Although, alternative experiments using a control part without an aptazyme sequence showed inhibitory effects of theophylline on the PURE cell-free system, the expression profiles and the cleavage characterization of BBa_K3806014, support that the lacZ expression from this part is ligand-responsive. For an improved version of this part, check BBa_K3806016.
References
- [1] Townshend, B., Xiang, J. S., Manzanarez, G., Hayden, E. J. and Smolke, C. (2021). A multiplexed, automated evolution pipeline enables scalable discovery and characterization of biosensors. Nat Commun, 12, 1437.
- [2] Klauser, B., & Hartig, J. S. (2013). An engineered small RNA-mediated genetic switch based on a ribozyme expression platform. Nucleic acids research, 41(10), 5542-5552.