Difference between revisions of "Part:BBa K3737004"
(→Improvement of ERE controlled TetR Expression - Alma 2021) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 8: | Line 8: | ||
| − | == | + | == Improvement of ERE controlled TetR Expression - Alma 2021 == |
| − | When we thought we had succeeded, we sent 2 colonies for sequencing and | + | When we thought we had succeeded in improving K123002, we sent 2 colonies for sequencing and learned that we had added a stronger promoter (Part BBa_J23100) and the human estrogen receptor element (Part BBa_K3737003). Although it showed that we were not able to include the ribosome binding site (RBS), we continued with qPCR to further compare parts K3737004 and K123002. |
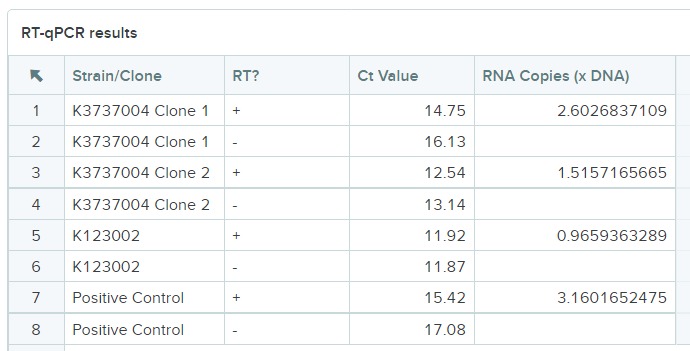
| − | This | + | When comparing K123002 to K3737004 via qPCR, we used the difference between CT values of the strains with and without reverse transcriptase (RT) to calculate the amount of RNA present. We used two clones of K3737004; both clones showed lower CT values with RT than without. This proves that more DNA was made from the RNA present in clones with RT compared to the clones without RT. |
| − | + | [[File:Alma qPCR.jpeg]] | |
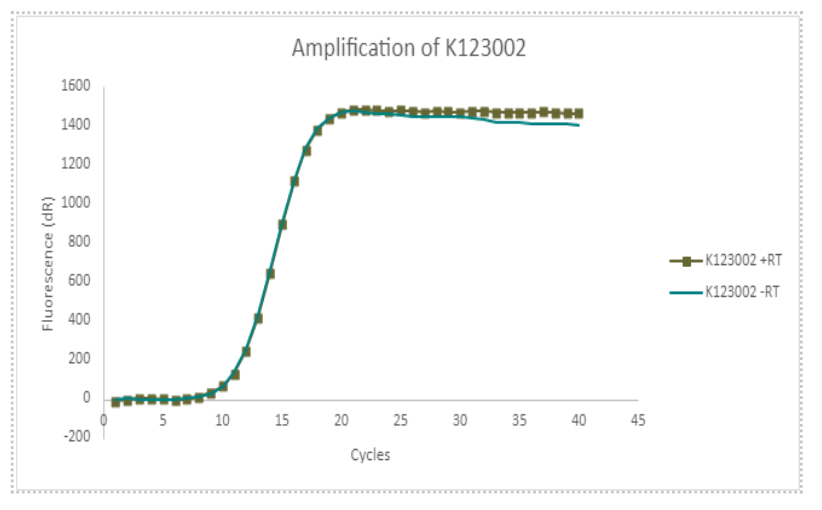
| − | + | K123002 had very similar CT values. This means that converting the RNA present did not make any more DNA, and there was no transcription of TetR in the K123002 part. Because we used tet_seqR and tet_seqF primers, we know that the TetR gene was present in K3737004 and not present in K123002. | |
| − | + | ||
| − | + | ||
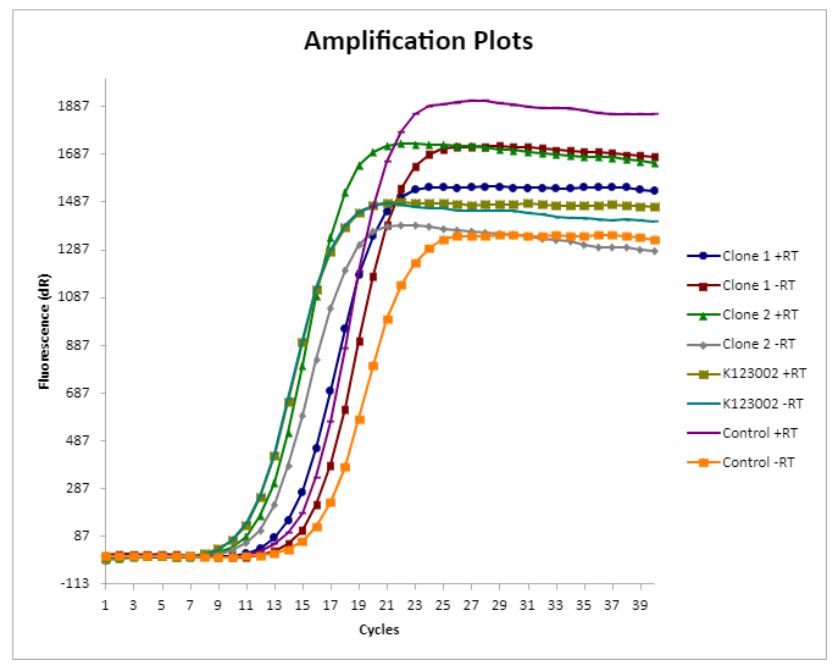
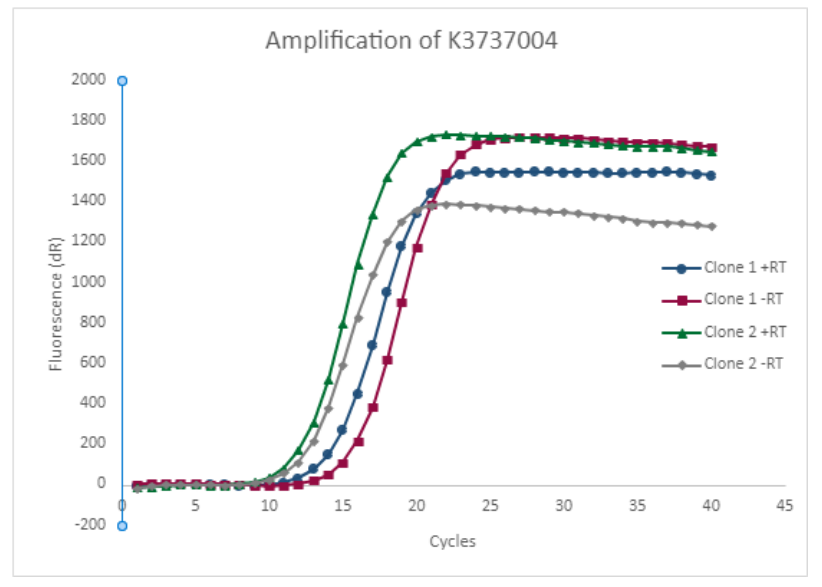
The amplification plot made from the qPCR is a visual representation of our data. We also made amplification plots to show the difference in amplification between K123002 and K3737004. | The amplification plot made from the qPCR is a visual representation of our data. We also made amplification plots to show the difference in amplification between K123002 and K3737004. | ||
| + | |||
| + | [[File:Alma qpcramplification.png]] | ||
| + | [[File:Alma amplificationK3737004.png]] | ||
| + | [[File:Alma amplificationK123002.png]] | ||
| + | |||
The dissociation curve shows only one peak, proving that there is only 1 type of DNA in the qPCR samples. This proves that the tet_seqR and tet_seqF were the only primers that were able to bind any genes in the samples. This is promising because it indicates that clone 1 and clone 2 are the same sequence. Based on this data, we have successfully improved upon K123002. | The dissociation curve shows only one peak, proving that there is only 1 type of DNA in the qPCR samples. This proves that the tet_seqR and tet_seqF were the only primers that were able to bind any genes in the samples. This is promising because it indicates that clone 1 and clone 2 are the same sequence. Based on this data, we have successfully improved upon K123002. | ||
| + | |||
| + | [[File:Alma qpcrdissociation]] | ||
<!-- --> | <!-- --> | ||
Latest revision as of 19:48, 21 October 2021
TetR repressor via estrogen binding
This part is an improvement upon the K123002 composite part. The improved part has a J23100 promoter in place of the LacIQ promoter and includes a ribosome binding site (RBS). The ssrA TetR degradation tag at the end of the DNA sequence was weakened.
This part is designed to be used with Estrogen Receptor (ER) because of its Human Estrogen Response Element (HERE). The ER binds the HERE and prevents TetR from getting promoted.
Improvement of ERE controlled TetR Expression - Alma 2021
When we thought we had succeeded in improving K123002, we sent 2 colonies for sequencing and learned that we had added a stronger promoter (Part BBa_J23100) and the human estrogen receptor element (Part BBa_K3737003). Although it showed that we were not able to include the ribosome binding site (RBS), we continued with qPCR to further compare parts K3737004 and K123002.
When comparing K123002 to K3737004 via qPCR, we used the difference between CT values of the strains with and without reverse transcriptase (RT) to calculate the amount of RNA present. We used two clones of K3737004; both clones showed lower CT values with RT than without. This proves that more DNA was made from the RNA present in clones with RT compared to the clones without RT.
K123002 had very similar CT values. This means that converting the RNA present did not make any more DNA, and there was no transcription of TetR in the K123002 part. Because we used tet_seqR and tet_seqF primers, we know that the TetR gene was present in K3737004 and not present in K123002.
The amplification plot made from the qPCR is a visual representation of our data. We also made amplification plots to show the difference in amplification between K123002 and K3737004.
The dissociation curve shows only one peak, proving that there is only 1 type of DNA in the qPCR samples. This proves that the tet_seqR and tet_seqF were the only primers that were able to bind any genes in the samples. This is promising because it indicates that clone 1 and clone 2 are the same sequence. Based on this data, we have successfully improved upon K123002.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]