Difference between revisions of "Part:BBa K3809012"
FCASTANEDA (Talk | contribs) |
|||
| Line 4: | Line 4: | ||
This sequence is the result of the assembly of two parts: BBa_K3809010 and BBa_K081005. The first one is the coding sequence of the Human Estrogen Receptor Alpha. It was modified in order to express the ligand binding domain and the DNA binding domain. The sequence also includes a 6X Histidine Tag (for its purification), a linker sequence of 4 glycines, 1 serine and 1 cistein (for its immobilization) and a signal peptide sequence (NPS4). The second part is a strong constitutive promoter with an RBS with an efficiency of 0.6. We used this part for the expression of the modified ESR1 protein and its subsequently purification so that we could use it for the detection of Endocrine Disrupting Chemicals or EDCs. | This sequence is the result of the assembly of two parts: BBa_K3809010 and BBa_K081005. The first one is the coding sequence of the Human Estrogen Receptor Alpha. It was modified in order to express the ligand binding domain and the DNA binding domain. The sequence also includes a 6X Histidine Tag (for its purification), a linker sequence of 4 glycines, 1 serine and 1 cistein (for its immobilization) and a signal peptide sequence (NPS4). The second part is a strong constitutive promoter with an RBS with an efficiency of 0.6. We used this part for the expression of the modified ESR1 protein and its subsequently purification so that we could use it for the detection of Endocrine Disrupting Chemicals or EDCs. | ||
| + | |||
| + | <div class=WordSection1> | ||
| + | |||
| + | <p class=MsoNormal><b><span lang=EN-US style='font-size:10.0pt;line-height: | ||
| + | 107%;font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'><o:p> </o:p></span></b></p> | ||
| + | |||
| + | <p class=MsoNormal><b><span lang=EN-US style='font-size:10.0pt;line-height: | ||
| + | 107%;font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'>Characterization of BBa_K3809012<o:p></o:p></span></b></p> | ||
| + | |||
| + | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'>For the characterization of the protein | ||
| + | BBa_K380912 we transformed the plasmid in <i>E. coli</i> BL21. We then examined | ||
| + | the growth of different colonies and performed a Colony PCR to determine if | ||
| + | they had the insert or not. The result can be seen in Figure 1. And 2. <span | ||
| + | class=SpellE>and</span> illustrates that only 6 out of 30 colonies we chose had | ||
| + | the insert. <o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><b style='mso-bidi-font-weight: | ||
| + | normal'><span style='font-size:15.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | color:#1155CC;mso-no-proof:yes'><img width=624 height=351 | ||
| + | src="https://2021.igem.org/wiki/images/2/27/T--TecCEM--PartsRegister2image002.jpg" | ||
| + | alt="Imagen que contiene Tabla Descripción generada automáticamente" | ||
| + | v:shapes="Imagen_x0020_3"><![endif]></span></b><span lang=EN-US | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'>Figure | ||
| + | 1.</span></b><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'> Gel electrophoresis of the colony PCR we | ||
| + | performed for the transformed bacteria BL21 from 1 to 19. We selected only | ||
| + | those colonies (marked in blue, M8, M13 and M18) that showed the insert at the | ||
| + | size we expected (1948 bp).<o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><b style='mso-bidi-font-weight: | ||
| + | normal'><span style='font-size:15.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | color:#1155CC;mso-no-proof:yes'><img width=624 height=351 | ||
| + | src="https://2021.igem.org/wiki/images/a/aa/T--TecCEM--PartsRegister2image004.jpg" | ||
| + | alt="Imagen que contiene Tabla Descripción generada automáticamente" | ||
| + | v:shapes="Imagen_x0020_4"><![endif]></span></b><span lang=EN-US | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'>Figure | ||
| + | 2.</span></b><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'> Gel electrophoresis of the colony PCR we | ||
| + | performed for the transformed bacteria BL21 from 20 to 30. We selected only | ||
| + | those colonies (marked in blue, M21, M23 and M26) that showed the insert at the | ||
| + | size we expected (1948 bp).<o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'><o:p> </o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'>After that, we propagated the selected | ||
| + | colonies on liquid LB+CAM medium and took samples of all the cultures at 1.5, | ||
| + | 3, 4.5 and 48 hours after their inoculation. We ran an SDS-PAGE electrophoresis | ||
| + | expecting to see a protein of around 69.5 <span class=SpellE>kDa</span> | ||
| + | (corresponding to the size of the protein receptor ESR1) but discovered that | ||
| + | only the colonies M18 and M23 had produced a visible protein at the expected site. | ||
| + | The best result corresponded to M23, which we describe below. <o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><span | ||
| + | style='font-family:"Arial",sans-serif;color:black;mso-no-proof:yes'><img width=545 height=268 | ||
| + | src="https://2021.igem.org/wiki/images/f/fa/T--TecCEM--PartsRegister2image006.jpg" v:shapes="Imagen_x0020_5"><![endif]></span><span | ||
| + | lang=EN-US style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'><o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal align=center style='text-align:center'><b><span lang=EN-US | ||
| + | style='font-size:10.0pt;line-height:107%;font-family:"Arial",sans-serif; | ||
| + | mso-fareast-font-family:"Times New Roman";color:black;mso-ansi-language:EN-US'>Figure | ||
| + | 3. </span></b><span lang=EN-US style='font-size:10.0pt;line-height:107%; | ||
| + | font-family:"Arial",sans-serif;mso-fareast-font-family:"Times New Roman"; | ||
| + | color:black;mso-ansi-language:EN-US'>SDS-PAGE electrophoresis showing the apparition | ||
| + | of a protein of around 70 <span class=SpellE>kDa</span> which corresponds to | ||
| + | ESR1.<o:p></o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal><span lang=EN-US style='mso-ansi-language:EN-US;mso-no-proof: | ||
| + | yes'><o:p> </o:p></span></p> | ||
| + | |||
| + | <p class=MsoNormal><span lang=EN-US style='mso-ansi-language:EN-US'><o:p> </o:p></span></p> | ||
| + | |||
| + | </div> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 16:09, 21 October 2021
Modified ESR1 protein under strong constitutive promoter and RBS
This sequence is the result of the assembly of two parts: BBa_K3809010 and BBa_K081005. The first one is the coding sequence of the Human Estrogen Receptor Alpha. It was modified in order to express the ligand binding domain and the DNA binding domain. The sequence also includes a 6X Histidine Tag (for its purification), a linker sequence of 4 glycines, 1 serine and 1 cistein (for its immobilization) and a signal peptide sequence (NPS4). The second part is a strong constitutive promoter with an RBS with an efficiency of 0.6. We used this part for the expression of the modified ESR1 protein and its subsequently purification so that we could use it for the detection of Endocrine Disrupting Chemicals or EDCs.
<o:p> </o:p>
Characterization of BBa_K3809012<o:p></o:p>
For the characterization of the protein BBa_K380912 we transformed the plasmid in E. coli BL21. We then examined the growth of different colonies and performed a Colony PCR to determine if they had the insert or not. The result can be seen in Figure 1. And 2. and illustrates that only 6 out of 30 colonies we chose had the insert. <o:p></o:p>
<img width=624 height=351
src="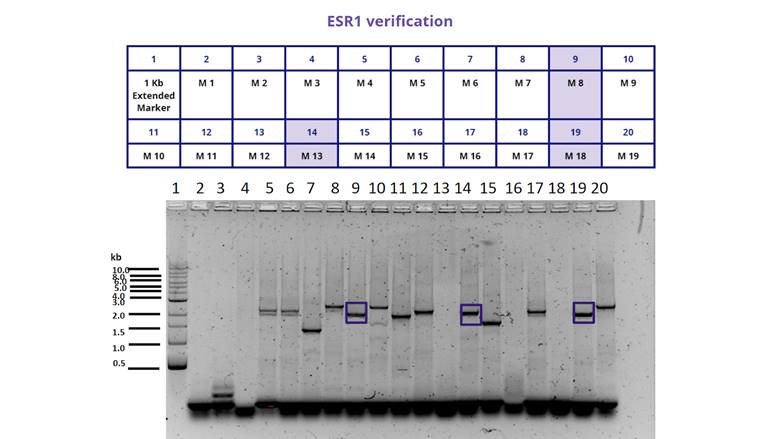 "
alt="Imagen que contiene Tabla
Descripción generada automáticamente"
v:shapes="Imagen_x0020_3"><![endif]><o:p></o:p>
"
alt="Imagen que contiene Tabla
Descripción generada automáticamente"
v:shapes="Imagen_x0020_3"><![endif]><o:p></o:p>
Figure 1. Gel electrophoresis of the colony PCR we performed for the transformed bacteria BL21 from 1 to 19. We selected only those colonies (marked in blue, M8, M13 and M18) that showed the insert at the size we expected (1948 bp).<o:p></o:p>
<img width=624 height=351
src=" "
alt="Imagen que contiene Tabla
Descripción generada automáticamente"
v:shapes="Imagen_x0020_4"><![endif]><o:p></o:p>
"
alt="Imagen que contiene Tabla
Descripción generada automáticamente"
v:shapes="Imagen_x0020_4"><![endif]><o:p></o:p>
Figure 2. Gel electrophoresis of the colony PCR we performed for the transformed bacteria BL21 from 20 to 30. We selected only those colonies (marked in blue, M21, M23 and M26) that showed the insert at the size we expected (1948 bp).<o:p></o:p>
<o:p> </o:p>
After that, we propagated the selected colonies on liquid LB+CAM medium and took samples of all the cultures at 1.5, 3, 4.5 and 48 hours after their inoculation. We ran an SDS-PAGE electrophoresis expecting to see a protein of around 69.5 kDa (corresponding to the size of the protein receptor ESR1) but discovered that only the colonies M18 and M23 had produced a visible protein at the expected site. The best result corresponded to M23, which we describe below. <o:p></o:p>
<img width=545 height=268
src=" " v:shapes="Imagen_x0020_5"><![endif]><o:p></o:p>
" v:shapes="Imagen_x0020_5"><![endif]><o:p></o:p>
Figure 3. SDS-PAGE electrophoresis showing the apparition of a protein of around 70 kDa which corresponds to ESR1.<o:p></o:p>
<o:p> </o:p>
<o:p> </o:p>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 112
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1328
