Difference between revisions of "Part:BBa K3825001"
Dragon210331 (Talk | contribs) (→Usage and Biology) |
|||
| Line 6: | Line 6: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ===CKWA-China 2021=== | |
This the coding sequence of Lon protease obtained from E. coli sources. Our project in 2021 uses this part in E. coli BL21(DE3), Rosetta and BL21(plus) strains to produce E. coli Lon protease. According to the reports in literatures, Lon protease decomposes C-Myc protein commonly found in cancer cells. Because of this potential, the protease is planned to be used to make medicine to suppress cancer recurrence. | This the coding sequence of Lon protease obtained from E. coli sources. Our project in 2021 uses this part in E. coli BL21(DE3), Rosetta and BL21(plus) strains to produce E. coli Lon protease. According to the reports in literatures, Lon protease decomposes C-Myc protein commonly found in cancer cells. Because of this potential, the protease is planned to be used to make medicine to suppress cancer recurrence. | ||
Revision as of 14:30, 21 October 2021
T7-LacO-rLon (E.coli)-6xHis
rLon (E.coli)-6xHis
Usage and Biology
CKWA-China 2021
This the coding sequence of Lon protease obtained from E. coli sources. Our project in 2021 uses this part in E. coli BL21(DE3), Rosetta and BL21(plus) strains to produce E. coli Lon protease. According to the reports in literatures, Lon protease decomposes C-Myc protein commonly found in cancer cells. Because of this potential, the protease is planned to be used to make medicine to suppress cancer recurrence.
We designed to use LacI and LacO together with T7 promoter to express this Lon protease in a commonly used plasmid pet28a(+).
Protocols
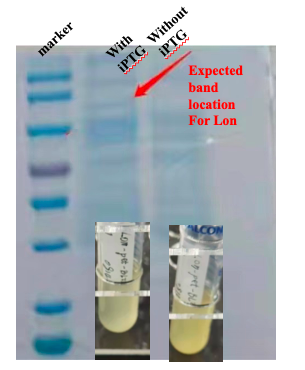
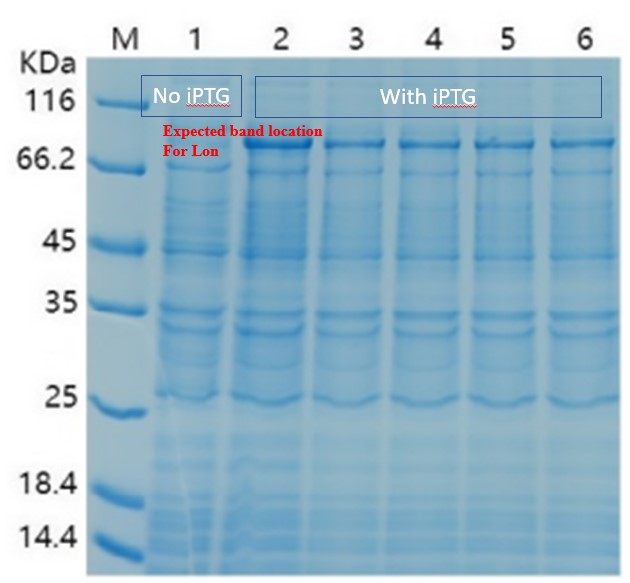
For the measurement, E. coli Rosetta containing the designed plasmid was incubated at 37C overnight with 1mM IPTG added. Then the cells are lysed by 200 μL 4% SDS for 10 minutes in room temperature, then 10 min at 95C. Then loading buffer is added. The SDS-PAGE was performed with a 12% precast polyacrylamide gel. Coomassie bright blue stain is used to show the bands.
With these induction conditions, in our first test we failed to observe a clear band of Lon protein. The bacteria culture was also relatively clear. It is possible that Lon protease has been decomposing the proteins needed for E. coli growth. Because the bacteria growth was suppressed by the Lon protease, its own production was low.
When there is LacI and there is LacO before the Lon gene sequence, IPTG can be used to control the expression. The control can be tuned by the concentration and timing of induction. We then changed our condition: the bacteria were cultured on LB plate overnight at 37C, and then pick a single colony and incubate in LB medium until the OD600 reached 0.6-0.8. At this time, IPTG was added at 0.5 mM, and the medium was again incubated at 37C for another 4 hours. This time the SDS-PAGE results showed remarkable expression of the Lon protease. The results confirmed the capability of this part, and also showed for better protein expression and better bateria growth, the timing for induction should also be considered and experimented.
Summary
Lon protease may have an impact on bacterial growth, and it is necessary to control appropriate induction reagents and culture conditions for better expression. We have initially found suitable conditions, and we can continue to optimize on this basis in the future to obtain higher output. And by extracting the protein, testing the function of the protein, we can determine the activity of the protein.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]