Difference between revisions of "Part:BBa K1483000"
EastonLiaw (Talk | contribs) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 16: | Line 16: | ||
<b><font size="+1.2">Improvement by 2021 TAS_Taipei</font></b> | <b><font size="+1.2">Improvement by 2021 TAS_Taipei</font></b> | ||
| − | We improved a part from the 2014 Tuebingen team: α-N-Acetylgalactosaminidase (NAGA) from | + | We improved a part from the 2014 Tuebingen team: α-N-Acetylgalactosaminidase (NAGA) from <i>Elizabethkingia meningoseptica</i>(Part:BBa_K1483000). |
| − | <b><font size="+0.5">Expression of Part From 2014 Tuebingen</font></b> | + | <b><font size="+0.5">Unsuccessful Expression of Part From 2014 Tuebingen</font></b> |
We obtained the amino acid sequence of the α-N-Acetylgalactosaminidase protein from the iGEM DNA Repository Plate (BBa_K1483000), as entered into the iGEM parts collection database by the Tuebingen iGEM team in 2014. | We obtained the amino acid sequence of the α-N-Acetylgalactosaminidase protein from the iGEM DNA Repository Plate (BBa_K1483000), as entered into the iGEM parts collection database by the Tuebingen iGEM team in 2014. | ||
| Line 26: | Line 26: | ||
https://static.igem.org/mediawiki/parts/5/55/T--TAS_Taipei--k88nagagelpic.jpg | https://static.igem.org/mediawiki/parts/5/55/T--TAS_Taipei--k88nagagelpic.jpg | ||
| − | <b>Figure 1 | + | <b>Figure 1. Colony PCR check for strong promoter (K88) α-N-Acetylgalactosaminidase (NAGA) (Part: BBa_K3717004) using VF2 and VR primers. Uncut plasmid (K88 only control) has a band at the expected part size of 355 bp, indicated by white triangle. Confirms successful ligation as a band is produced at the expected size of 1684 bp, as indicated by the red triangle.</b> |
https://2021.igem.org/wiki/images/9/94/T--TAS_Taipei--engineeringsucess2.png | https://2021.igem.org/wiki/images/9/94/T--TAS_Taipei--engineeringsucess2.png | ||
| − | <b>Figure 2 | + | <b>Figure 2. Colony PCR check for T7 promoter α-N-Acetylgalactosaminidase (NAGA) (Part: BBa_K3717002) using VF2 and VR primers. Uncut plasmid (T7 only control) has a band at the expected part size of 332bp, indicated by white triangle. Confirms successful ligation as a band is produced at the expected size of 1661bp, indicated by the red triangle.</b> |
| − | We tested protein expression of these two composite parts by transforming our plasmids into BL21 E. | + | We tested protein expression of these two composite parts by transforming our plasmids into BL21 <i>E. coli</i> cells. We grew cultures at 37°C overnight, diluted those cultures, and then grew to OD600 0.5~0.6 at 37°C. We then induced expression with 0.5 mM IPTG and allowed cultures to grow overnight at room temperature. We took samples pre-induction and post-induction and examined them by SDS-PAGE. |
https://2021.igem.org/wiki/images/6/6b/T--TAS_Taipei--engineeringsucess3.png | https://2021.igem.org/wiki/images/6/6b/T--TAS_Taipei--engineeringsucess3.png | ||
| − | <b>Figure 3 | + | <b>Figure 3. SDS-PAGE of cell lysate for each strain: T7 promoter α-N-Acetylgalactosaminidase (NAGA) and strong promoter (K88) α-N-Acetylgalactosaminidase (NAGA). Blue triangles indicate expected size for NAGA (50.1 kDa). Sequences for target proteins do not contain a start codon, thus have no expression, as shown by the triangles. </b> |
Our SDS-page did not show any overexpression bands for the enzymes of interest. The results indicate that there were no target proteins at their expected band sizes: 50.1 kDa band for both T7 promoter + NAGA and K88 promoter + NAGA in the induced sample. As the SDS page is of cell lysis samples, other bands present are due to innate proteins present in the bacteria cell. | Our SDS-page did not show any overexpression bands for the enzymes of interest. The results indicate that there were no target proteins at their expected band sizes: 50.1 kDa band for both T7 promoter + NAGA and K88 promoter + NAGA in the induced sample. As the SDS page is of cell lysis samples, other bands present are due to innate proteins present in the bacteria cell. | ||
| − | Upon comparison of the amino acid sequence from Tuebingen’s part (BBa_K1483000) with full sequences that were offered by other studies online, we discovered that the enzyme sequences were missing the start codon (Fig | + | Upon comparison of the amino acid sequence from Tuebingen’s part (BBa_K1483000) with full sequences that were offered by other studies online, we discovered that the enzyme sequences were missing the start codon (Fig 4), which explained the non-expression of the proteins. |
https://static.igem.org/mediawiki/parts/2/2d/T--TAS_Taipei--nagaaminoacids.jpg | https://static.igem.org/mediawiki/parts/2/2d/T--TAS_Taipei--nagaaminoacids.jpg | ||
| − | <b>Figure 4 | + | <b>Figure 4. Top sequence: First 37 amino acids of Team Tuebingen's 2014</b> |
α-N-Acetylgalactosaminidase part BBa_K1483000. Bottom sequence: First 38 amino acids of TAS_Taipei's α-N-Acetylgalactosaminidase part BBa_K3717016. Based on the alignment of the two sequences, Tuebingen's part is missing the first amino acid of the α-N-Acetylgalactosaminidase protein. | α-N-Acetylgalactosaminidase part BBa_K1483000. Bottom sequence: First 38 amino acids of TAS_Taipei's α-N-Acetylgalactosaminidase part BBa_K3717016. Based on the alignment of the two sequences, Tuebingen's part is missing the first amino acid of the α-N-Acetylgalactosaminidase protein. | ||
| Line 55: | Line 55: | ||
<b><font size="+0.5">Successful Expression of New Improved Part</font></b> | <b><font size="+0.5">Successful Expression of New Improved Part</font></b> | ||
| − | BBa_K3717016 was added to the iGEM parts registry. We derived the sequence of α-N-Acetylgalactosaminidase (NAGA) from Elizabethkingia meningoseptica | + | BBa_K3717016 was added to the iGEM parts registry. We derived the sequence of α-N-Acetylgalactosaminidase (NAGA) from <i>Elizabethkingia meningoseptica</i> [1] then codon optimized the sequence for <i>E. coli</i> protein expression and attached a 6x Histidine tag (His-tag) downstream of the protein sequence through a glycine-serine linker (GS linker). This formed a new α-N-Acetylgalactosaminidase basic part with a C-Terminal His-tag downstream of the protein sequence (BBa_K3717016). |
https://static.igem.org/mediawiki/parts/c/ca/T--TAS_Taipei--nagahis.jpg | https://static.igem.org/mediawiki/parts/c/ca/T--TAS_Taipei--nagahis.jpg | ||
| − | <b>Figure | + | <b>Figure 5. Open reading frame for α-N-Acetylgalactosaminidase with C-Terminal 6x Histidine tag (BBa_K3717016)</b> |
| − | NAGA is an enzyme that catalyzes the cleavage of the N-acetylgalactosamine off of A type blood antigens such that the remaining sugar can be classified as an H antigen which the anti-A and anti-B antibodies are unable to recognize and hence does not elicit an immune response in the human body | + | NAGA is an enzyme that catalyzes the cleavage of the N-acetylgalactosamine off of A type blood antigens such that the remaining sugar can be classified as an H antigen which the anti-A and anti-B antibodies are unable to recognize and hence does not elicit an immune response in the human body [2]. Thus, NAGA can convert A blood types to universal O type. |
In order to test our sequence, we attached a T7 promoter + RBS (BBa_K525998) upstream of the open reading frame (ORF) and double terminator (BBa_B0015) downstream of the ORF, which formed our composite part (BBa_K3717013). | In order to test our sequence, we attached a T7 promoter + RBS (BBa_K525998) upstream of the open reading frame (ORF) and double terminator (BBa_B0015) downstream of the ORF, which formed our composite part (BBa_K3717013). | ||
| Line 67: | Line 67: | ||
https://static.igem.org/mediawiki/parts/f/f5/T--TAS_Taipei--t7nagahis.jpg | https://static.igem.org/mediawiki/parts/f/f5/T--TAS_Taipei--t7nagahis.jpg | ||
| − | <b>Figure | + | <b>Figure 6. Design of α-N-Acetylgalactosaminidase with T7 Promoter, strong RBS, C-Terminal 6x Histidine tag and Double Terminator Construct (BBa_K3717013)</b> |
| − | We tested protein expression of | + | We tested protein expression of the composite parts by transforming our plasmids into BL21 <i>E. coli</i> cells. We grew cultures at 37°C overnight, diluted those cultures, and then grew to OD600 0.5~0.6 at 37°C. We then induced expression with 0.5 mM IPTG and allowed cultures to grow overnight at room temperature. We harvested cells by centrifugation and lysed cell pellets through either sonication or with xTractor Lysis Buffer [3] supplemented with 20 mM imidazole. We purified our histidine-tagged proteins using Ni sepharose affinity chromatography. We then utilized SDS-PAGE to confirm the sizes of purified proteins. |
https://2021.igem.org/wiki/images/b/ba/T--TAS_Taipei--engineeringsucess6.png | https://2021.igem.org/wiki/images/b/ba/T--TAS_Taipei--engineeringsucess6.png | ||
| − | <b>Figure | + | <b>Figure 7. SDS-PAGE of purified proteins with the T7 promoter α-N-Acetylgalactosaminidase expressing construct (BBa_K3717013). Band matches expected size of 51.7 kDa, proving successful expression and purification. </b> |
| + | |||
| + | Results from the gel (Fig. 7) show a band expressed at an expected size of 51.7 kDa for our new α-N-Acetylgalactosaminidase constructs, while no band expressed at an expected size of 51.7 kDa for Tuebingen’s α-N-Acetylgalactosaminidase constructs. This confirms our improvement of Tuebingen’s part BBa_K1483000 that was unable to express due to a lack of a start codon. | ||
| − | |||
| Line 84: | Line 85: | ||
We confirmed the functionality of purified NAGA by using two assays: colorimetric tests and mass spectroscopy. | We confirmed the functionality of purified NAGA by using two assays: colorimetric tests and mass spectroscopy. | ||
| − | We obtained a colorimetric substrate for NAGA from Sigma Aldrich (Fig | + | We obtained a colorimetric substrate for NAGA from Sigma Aldrich (Fig 8). This substrate contains a 4-nitrophenol leaving group, which turns yellow upon successful cleavage in solution. The concentration of 4-nitrophenol was quantified at absorbance 405 nm using a 96-well plate-based assay [4]. |
https://2021.igem.org/wiki/images/e/e6/T--TAS_Taipei--engineeringsucess7.png | https://2021.igem.org/wiki/images/e/e6/T--TAS_Taipei--engineeringsucess7.png | ||
| − | <b>Figure | + | <b>Figure 8. Colorimetric substrates for NAGA.</b> |
| − | We performed small-scale colorimetric tests to verify the function of our purified enzymes. To each well, we added 50 μL of 10 mM substrate, 10 μL of enzyme, 30 μL of water, and 10 μL of 10x Glycobuffer 1 (50 mM CaCl2, 500 mM sodium acetate, pH 5.5), a buffer recommended by New England Biolabs to ensure optimal enzyme activity | + | We performed small-scale colorimetric tests to verify the function of our purified enzymes. To each well, we added 50 μL of 10 mM substrate, 10 μL of enzyme, 30 μL of water, and 10 μL of 10x Glycobuffer 1 (50 mM CaCl2, 500 mM sodium acetate, pH 5.5), a buffer recommended by New England Biolabs to ensure optimal enzyme activity [5]. Following 2 hours of reaction at room temperature, we diluted the well contents in 1.9 mL of water and took absorbance readings at 405 nm. Our results indicated that NAGA successfully cleaved the substrates (Fig 9). Moreover, we demonstrated the specificity of the enzyme as it did not cleave the substrate for other enzymes. |
https://static.igem.org/mediawiki/parts/a/a6/T--TAS_Taipei--colorimetricnumerodos.png | https://static.igem.org/mediawiki/parts/a/a6/T--TAS_Taipei--colorimetricnumerodos.png | ||
| − | <b>Figure | + | <b>Figure 9. Absorbance of reaction solutions in small-scale colorimetric tests verify the functionality and specificity of the enzymes. a) 96-well plates following enzyme reaction. b) absorbance readings of diluted well contents (accounted for dilution). c) buffers and solutions used in substrate and control wells. </b> |
| − | To quantify the activity of NAGA, we performed enzyme reactions at various substrate concentrations and measured the absorbance at different time intervals over a constant time period (Fig | + | To quantify the activity of NAGA, we performed enzyme reactions at various substrate concentrations and measured the absorbance at different time intervals over a constant time period (Fig 10). We used the averages from at least three independent trials to calculate the Michaelis–Menten constant for enzyme efficiency (see enzyme model in modeling). The results of our colorimetric substrate tests show that NAGA enzyme is specific and functions as expected. |
https://2021.igem.org/wiki/images/5/55/T--TAS_Taipei--n.png | https://2021.igem.org/wiki/images/5/55/T--TAS_Taipei--n.png | ||
| − | <b>Figure | + | <b>Figure 10. Absorbance over time for various NAGA substrate concentrations</b> |
<b><font size="+0.2">Mass Spectroscopy Trisaccharide Antigen Tests</font></b> | <b><font size="+0.2">Mass Spectroscopy Trisaccharide Antigen Tests</font></b> | ||
| − | Given that the colorimetric results were positive, we wanted to further test the enzyme’s specificity by using trisaccharides as substrates. These trisaccharides contain three monosaccharides that have identical chemical structures to the A and B antigens found on RBCs. To test a proof of concept functionality of NAGA in cleaving the A-antigen trisaccharide, we carried out a reaction of the enzyme and its trisaccharide dissolved in 1X GlycoBuffer | + | Given that the colorimetric results were positive, we wanted to further test the enzyme’s specificity by using trisaccharides as substrates. These trisaccharides contain three monosaccharides that have identical chemical structures to the A and B antigens found on RBCs. To test a proof of concept functionality of NAGA in cleaving the A-antigen trisaccharide, we carried out a reaction of the enzyme and its trisaccharide dissolved in 1X GlycoBuffer [5], deionized water, and purified BSA for 1 hour at 37°C. The A-antigen trisaccharides were kindly supplied by Professor Dr. Todd Lowary from the Institute of Biological Chemistry at Academic Sinica [6]. |
For the enzyme tests, a non-specific trisaccharide served as a negative control and A-antigen trisaccharide specific to NAGA served as the experimental unit (Table 1). | For the enzyme tests, a non-specific trisaccharide served as a negative control and A-antigen trisaccharide specific to NAGA served as the experimental unit (Table 1). | ||
| Line 113: | Line 114: | ||
https://static.igem.org/mediawiki/parts/e/eb/T--TAS_Taipei--table3.png | https://static.igem.org/mediawiki/parts/e/eb/T--TAS_Taipei--table3.png | ||
| − | After the reaction, the reaction solution was passed through C18 columns to minimize impurities from the reaction solution. We then evaluated the flow through solution with a mass spectrometer to measure peaks in molar mass in order to determine the compounds present in the reaction solution. The original molar mass of the A-antigen is 713.35 g/mol; therefore, after cleavage, we expected two fragments with molar masses of 510.27 g/mol and 203.08 g/mol to form. (Fig | + | After the reaction, the reaction solution was passed through C18 columns to minimize impurities from the reaction solution. We then evaluated the flow through solution with a mass spectrometer to measure peaks in molar mass in order to determine the compounds present in the reaction solution. The original molar mass of the A-antigen is 713.35 g/mol; therefore, after cleavage, we expected two fragments with molar masses of 510.27 g/mol and 203.08 g/mol to form. (Fig 11). |
https://2021.igem.org/wiki/images/0/03/T--TAS_Taipei--nmass.png | https://2021.igem.org/wiki/images/0/03/T--TAS_Taipei--nmass.png | ||
| − | <b>Figure | + | <b>Figure 11. Molecular structure and mass of A-antigen trisaccharides and subsequent products following enzymatic cleavage. </b> |
| − | The results shown by the mass spectrometer confirm the functionality of NAGA. The experimental unit shows the presence of the cleaved fragment from the reaction with A-antigen trisaccharide, indicated by the peak at 533 g/mol in the mass spectrum (Fig | + | The results shown by the mass spectrometer confirm the functionality of NAGA. The experimental unit shows the presence of the cleaved fragment from the reaction with A-antigen trisaccharide, indicated by the peak at 533 g/mol in the mass spectrum (Fig 12). The negative control confirms the specificity of NAGA, since it did not cleave the B-antigen trisaccharide, as indicated by the peak at 695 g/mol (Fig 13). Throughout all readings of the mass spectrum, the molar mass of the peaks have been increased by 23 g/mol due to a sodium adduct. Unrelated peaks/background noise can be attributed to compounds present in the buffer solution. |
https://static.igem.org/mediawiki/parts/9/9d/T--TAS_Taipei--newimage3.png | https://static.igem.org/mediawiki/parts/9/9d/T--TAS_Taipei--newimage3.png | ||
| − | <b>Figure | + | <b>Figure 12. α-N-Acetylgalactosaminidase cleaves A-antigen trisaccharide. Mass spectrum of α-N-Acetylgalactosaminidase and A-antigen trisaccharide reaction solution (experimental unit, flow through). The peak at 533g/mol shows the presence of the cleaved fragment from the reaction of the α-N-Acetylgalactosaminidase enzyme and A-antigen trisaccharide.</b> |
https://static.igem.org/mediawiki/parts/1/14/T--TAS_Taipei--newimage4.png | https://static.igem.org/mediawiki/parts/1/14/T--TAS_Taipei--newimage4.png | ||
| − | <b>Figure | + | <b>Figure 13. Negative Control: Mass spectrum of α-N-Acetylgalactosaminidase and B-antigen trisaccharide reaction solution (negative control, flow through). The negative control confirms the specificity of α-N-Acetylgalactosaminidase, as the B-antigen trisaccharide was not cleaved, indicated by the peak at 695g/mol</b> |
| − | Mass spectroscopy results from the | + | Mass spectroscopy results from the reaction between NAGA and A-antigen trisaccharide substrate demonstrate its ability to cleave its specific trisaccharides, demonstrating the proof of concept that the enzyme is able to cleave the A blood group antigens. |
| Line 137: | Line 138: | ||
<b>References</b> | <b>References</b> | ||
| − | + | 1. UniProtKB - A4Q8F7 (GH109_ELIME). UniProt, 2 June 2021, www.uniprot.org/uniprot/A4Q8F7. Accessed 20 Oct. 2021. | |
| − | XTractorTM Buffer & xTractor Buffer Kit User Manual. (n.d.). 10. | + | 2. Rahfeld, Peter, and Stephen G. Withers. “Toward Universal Donor Blood: Enzymatic Conversion of A and B to O Type.” Journal of Biological Chemistry, vol. 295, no. 2, Jan. 2020, pp. 325–34. DOI.org (Crossref), https://doi.org/10.1074/jbc.REV119.008164. |
| + | |||
| + | 3. XTractorTM Buffer & xTractor Buffer Kit User Manual. (n.d.). 10. | ||
Held, Paul. “Kinetic Analysis of β-Galactosidase Activity Using the PowerWave™ HT and Gen5™ Data Analysis Software .” BioTek, 16 Feb. 2007. | Held, Paul. “Kinetic Analysis of β-Galactosidase Activity Using the PowerWave™ HT and Gen5™ Data Analysis Software .” BioTek, 16 Feb. 2007. | ||
| − | + | 4. Held, Paul. “Kinetic Analysis of β-Galactosidase Activity Using the PowerWave™ HT and Gen5™ Data Analysis Software .” BioTek, 16 Feb. 2007. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| + | 5. Biolabs, New England. “Typical Reaction Conditions for α-N-Acetylgalactosaminidase (P0734).” New England Biolabs: Reagents for the Life Sciences Industry, https://international.neb.com/protocols/2013/01/10/typical-reaction-conditions-p0734. | ||
| + | 6. Meloncelli, Peter J., and Todd L. Lowary. “Synthesis of Abo Histo-Blood Group Type I and II Antigens.” Carbohydrate Research, vol. 345, no. 16, 16 Sept. 2010, pp. 2305–2322., https://doi.org/10.1016/j.carres.2010.08.012. | ||
Latest revision as of 12:03, 21 October 2021
α-N-Acetylgalactosamindase
This part is [http://appft1.uspto.gov/netacgi/nph-Parser?Sect1=PTO1&Sect2=HITOFF&d=PG01&p=1&u=/netahtml/PTO/srchnum.html&r=1&f=G&l=50&s1=20140220553.PGNR. patented] and should therfore only be used for research purposes.
Encodes for an enzyme, that is capable of cleaving off N-acetlygalactosamine from A-group blood antigens, thereby turning them into H-antigens. Can be used to convert erythrocytes from blood type A to O. Part in RFC25 standard.
Following reaction is catalysed by this part
Improvement by 2021 TAS_Taipei
We improved a part from the 2014 Tuebingen team: α-N-Acetylgalactosaminidase (NAGA) from Elizabethkingia meningoseptica(Part:BBa_K1483000).
Unsuccessful Expression of Part From 2014 Tuebingen
We obtained the amino acid sequence of the α-N-Acetylgalactosaminidase protein from the iGEM DNA Repository Plate (BBa_K1483000), as entered into the iGEM parts collection database by the Tuebingen iGEM team in 2014.
In order to test protein expression of the enzyme, we added a strong promoter and strong ribosome binding site (RBS; BBa_K880005) upstream of the protein amino acid sequence to create a part BBa_K3717004, as well as a T7 promoter and strong ribosome binding site (RBS; BBa_K525998) upstream of the protein amino acid sequence to create a part BBa_K3717002.

Figure 1. Colony PCR check for strong promoter (K88) α-N-Acetylgalactosaminidase (NAGA) (Part: BBa_K3717004) using VF2 and VR primers. Uncut plasmid (K88 only control) has a band at the expected part size of 355 bp, indicated by white triangle. Confirms successful ligation as a band is produced at the expected size of 1684 bp, as indicated by the red triangle.

Figure 2. Colony PCR check for T7 promoter α-N-Acetylgalactosaminidase (NAGA) (Part: BBa_K3717002) using VF2 and VR primers. Uncut plasmid (T7 only control) has a band at the expected part size of 332bp, indicated by white triangle. Confirms successful ligation as a band is produced at the expected size of 1661bp, indicated by the red triangle.
We tested protein expression of these two composite parts by transforming our plasmids into BL21 E. coli cells. We grew cultures at 37°C overnight, diluted those cultures, and then grew to OD600 0.5~0.6 at 37°C. We then induced expression with 0.5 mM IPTG and allowed cultures to grow overnight at room temperature. We took samples pre-induction and post-induction and examined them by SDS-PAGE.

Figure 3. SDS-PAGE of cell lysate for each strain: T7 promoter α-N-Acetylgalactosaminidase (NAGA) and strong promoter (K88) α-N-Acetylgalactosaminidase (NAGA). Blue triangles indicate expected size for NAGA (50.1 kDa). Sequences for target proteins do not contain a start codon, thus have no expression, as shown by the triangles.
Our SDS-page did not show any overexpression bands for the enzymes of interest. The results indicate that there were no target proteins at their expected band sizes: 50.1 kDa band for both T7 promoter + NAGA and K88 promoter + NAGA in the induced sample. As the SDS page is of cell lysis samples, other bands present are due to innate proteins present in the bacteria cell.
Upon comparison of the amino acid sequence from Tuebingen’s part (BBa_K1483000) with full sequences that were offered by other studies online, we discovered that the enzyme sequences were missing the start codon (Fig 4), which explained the non-expression of the proteins.

Figure 4. Top sequence: First 37 amino acids of Team Tuebingen's 2014
α-N-Acetylgalactosaminidase part BBa_K1483000. Bottom sequence: First 38 amino acids of TAS_Taipei's α-N-Acetylgalactosaminidase part BBa_K3717016. Based on the alignment of the two sequences, Tuebingen's part is missing the first amino acid of the α-N-Acetylgalactosaminidase protein.
Successful Expression of New Improved Part
BBa_K3717016 was added to the iGEM parts registry. We derived the sequence of α-N-Acetylgalactosaminidase (NAGA) from Elizabethkingia meningoseptica [1] then codon optimized the sequence for E. coli protein expression and attached a 6x Histidine tag (His-tag) downstream of the protein sequence through a glycine-serine linker (GS linker). This formed a new α-N-Acetylgalactosaminidase basic part with a C-Terminal His-tag downstream of the protein sequence (BBa_K3717016).

Figure 5. Open reading frame for α-N-Acetylgalactosaminidase with C-Terminal 6x Histidine tag (BBa_K3717016)
NAGA is an enzyme that catalyzes the cleavage of the N-acetylgalactosamine off of A type blood antigens such that the remaining sugar can be classified as an H antigen which the anti-A and anti-B antibodies are unable to recognize and hence does not elicit an immune response in the human body [2]. Thus, NAGA can convert A blood types to universal O type.
In order to test our sequence, we attached a T7 promoter + RBS (BBa_K525998) upstream of the open reading frame (ORF) and double terminator (BBa_B0015) downstream of the ORF, which formed our composite part (BBa_K3717013).
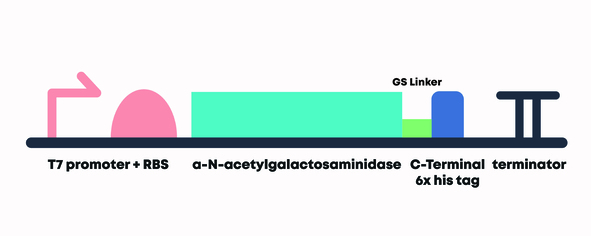
Figure 6. Design of α-N-Acetylgalactosaminidase with T7 Promoter, strong RBS, C-Terminal 6x Histidine tag and Double Terminator Construct (BBa_K3717013)
We tested protein expression of the composite parts by transforming our plasmids into BL21 E. coli cells. We grew cultures at 37°C overnight, diluted those cultures, and then grew to OD600 0.5~0.6 at 37°C. We then induced expression with 0.5 mM IPTG and allowed cultures to grow overnight at room temperature. We harvested cells by centrifugation and lysed cell pellets through either sonication or with xTractor Lysis Buffer [3] supplemented with 20 mM imidazole. We purified our histidine-tagged proteins using Ni sepharose affinity chromatography. We then utilized SDS-PAGE to confirm the sizes of purified proteins.

Figure 7. SDS-PAGE of purified proteins with the T7 promoter α-N-Acetylgalactosaminidase expressing construct (BBa_K3717013). Band matches expected size of 51.7 kDa, proving successful expression and purification.
Results from the gel (Fig. 7) show a band expressed at an expected size of 51.7 kDa for our new α-N-Acetylgalactosaminidase constructs, while no band expressed at an expected size of 51.7 kDa for Tuebingen’s α-N-Acetylgalactosaminidase constructs. This confirms our improvement of Tuebingen’s part BBa_K1483000 that was unable to express due to a lack of a start codon.
Successful Function of New Improved Part
Colorimetric Substrate Tests
We confirmed the functionality of purified NAGA by using two assays: colorimetric tests and mass spectroscopy.
We obtained a colorimetric substrate for NAGA from Sigma Aldrich (Fig 8). This substrate contains a 4-nitrophenol leaving group, which turns yellow upon successful cleavage in solution. The concentration of 4-nitrophenol was quantified at absorbance 405 nm using a 96-well plate-based assay [4].

Figure 8. Colorimetric substrates for NAGA.
We performed small-scale colorimetric tests to verify the function of our purified enzymes. To each well, we added 50 μL of 10 mM substrate, 10 μL of enzyme, 30 μL of water, and 10 μL of 10x Glycobuffer 1 (50 mM CaCl2, 500 mM sodium acetate, pH 5.5), a buffer recommended by New England Biolabs to ensure optimal enzyme activity [5]. Following 2 hours of reaction at room temperature, we diluted the well contents in 1.9 mL of water and took absorbance readings at 405 nm. Our results indicated that NAGA successfully cleaved the substrates (Fig 9). Moreover, we demonstrated the specificity of the enzyme as it did not cleave the substrate for other enzymes.

Figure 9. Absorbance of reaction solutions in small-scale colorimetric tests verify the functionality and specificity of the enzymes. a) 96-well plates following enzyme reaction. b) absorbance readings of diluted well contents (accounted for dilution). c) buffers and solutions used in substrate and control wells.
To quantify the activity of NAGA, we performed enzyme reactions at various substrate concentrations and measured the absorbance at different time intervals over a constant time period (Fig 10). We used the averages from at least three independent trials to calculate the Michaelis–Menten constant for enzyme efficiency (see enzyme model in modeling). The results of our colorimetric substrate tests show that NAGA enzyme is specific and functions as expected.

Figure 10. Absorbance over time for various NAGA substrate concentrations
Mass Spectroscopy Trisaccharide Antigen Tests
Given that the colorimetric results were positive, we wanted to further test the enzyme’s specificity by using trisaccharides as substrates. These trisaccharides contain three monosaccharides that have identical chemical structures to the A and B antigens found on RBCs. To test a proof of concept functionality of NAGA in cleaving the A-antigen trisaccharide, we carried out a reaction of the enzyme and its trisaccharide dissolved in 1X GlycoBuffer [5], deionized water, and purified BSA for 1 hour at 37°C. The A-antigen trisaccharides were kindly supplied by Professor Dr. Todd Lowary from the Institute of Biological Chemistry at Academic Sinica [6].
For the enzyme tests, a non-specific trisaccharide served as a negative control and A-antigen trisaccharide specific to NAGA served as the experimental unit (Table 1).
Table 1. Experimental setup for enzyme-antigen tests.

After the reaction, the reaction solution was passed through C18 columns to minimize impurities from the reaction solution. We then evaluated the flow through solution with a mass spectrometer to measure peaks in molar mass in order to determine the compounds present in the reaction solution. The original molar mass of the A-antigen is 713.35 g/mol; therefore, after cleavage, we expected two fragments with molar masses of 510.27 g/mol and 203.08 g/mol to form. (Fig 11).

Figure 11. Molecular structure and mass of A-antigen trisaccharides and subsequent products following enzymatic cleavage.
The results shown by the mass spectrometer confirm the functionality of NAGA. The experimental unit shows the presence of the cleaved fragment from the reaction with A-antigen trisaccharide, indicated by the peak at 533 g/mol in the mass spectrum (Fig 12). The negative control confirms the specificity of NAGA, since it did not cleave the B-antigen trisaccharide, as indicated by the peak at 695 g/mol (Fig 13). Throughout all readings of the mass spectrum, the molar mass of the peaks have been increased by 23 g/mol due to a sodium adduct. Unrelated peaks/background noise can be attributed to compounds present in the buffer solution.

Figure 12. α-N-Acetylgalactosaminidase cleaves A-antigen trisaccharide. Mass spectrum of α-N-Acetylgalactosaminidase and A-antigen trisaccharide reaction solution (experimental unit, flow through). The peak at 533g/mol shows the presence of the cleaved fragment from the reaction of the α-N-Acetylgalactosaminidase enzyme and A-antigen trisaccharide.

Figure 13. Negative Control: Mass spectrum of α-N-Acetylgalactosaminidase and B-antigen trisaccharide reaction solution (negative control, flow through). The negative control confirms the specificity of α-N-Acetylgalactosaminidase, as the B-antigen trisaccharide was not cleaved, indicated by the peak at 695g/mol
Mass spectroscopy results from the reaction between NAGA and A-antigen trisaccharide substrate demonstrate its ability to cleave its specific trisaccharides, demonstrating the proof of concept that the enzyme is able to cleave the A blood group antigens.
References
1. UniProtKB - A4Q8F7 (GH109_ELIME). UniProt, 2 June 2021, www.uniprot.org/uniprot/A4Q8F7. Accessed 20 Oct. 2021.
2. Rahfeld, Peter, and Stephen G. Withers. “Toward Universal Donor Blood: Enzymatic Conversion of A and B to O Type.” Journal of Biological Chemistry, vol. 295, no. 2, Jan. 2020, pp. 325–34. DOI.org (Crossref), https://doi.org/10.1074/jbc.REV119.008164.
3. XTractorTM Buffer & xTractor Buffer Kit User Manual. (n.d.). 10. Held, Paul. “Kinetic Analysis of β-Galactosidase Activity Using the PowerWave™ HT and Gen5™ Data Analysis Software .” BioTek, 16 Feb. 2007.
4. Held, Paul. “Kinetic Analysis of β-Galactosidase Activity Using the PowerWave™ HT and Gen5™ Data Analysis Software .” BioTek, 16 Feb. 2007.
5. Biolabs, New England. “Typical Reaction Conditions for α-N-Acetylgalactosaminidase (P0734).” New England Biolabs: Reagents for the Life Sciences Industry, https://international.neb.com/protocols/2013/01/10/typical-reaction-conditions-p0734.
6. Meloncelli, Peter J., and Todd L. Lowary. “Synthesis of Abo Histo-Blood Group Type I and II Antigens.” Carbohydrate Research, vol. 345, no. 16, 16 Sept. 2010, pp. 2305–2322., https://doi.org/10.1016/j.carres.2010.08.012.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 405

