Difference between revisions of "Part:BBa K398108"
Ravandervalk (Talk | contribs) (→Results) |
Ananya b1211 (Talk | contribs) |
||
| (22 intermediate revisions by 5 users not shown) | |||
| Line 10: | Line 10: | ||
[[Image:TU_Delft_Salt_tolerance.jpg|500px|right]] | [[Image:TU_Delft_Salt_tolerance.jpg|500px|right]] | ||
The salt tolerance induces by this BioBrick was tested by using NaCl in LB and M9 (containing 40 mM casaminoacids) media from 0 to 1M salt concentrations, it was compared to another of the TU Delft 2010 BioBricks which had the same antibiotic resistance, and same expression level (but of another protein which did not cause increased salt tolerance). | The salt tolerance induces by this BioBrick was tested by using NaCl in LB and M9 (containing 40 mM casaminoacids) media from 0 to 1M salt concentrations, it was compared to another of the TU Delft 2010 BioBricks which had the same antibiotic resistance, and same expression level (but of another protein which did not cause increased salt tolerance). | ||
| − | + | abc | |
The [http://2010.igem.org/Team:TU_Delft#page=Project/tolerance characterization of BBa_K398108] has been described on the TU Delft iGEM Team 2010 wiki. | The [http://2010.igem.org/Team:TU_Delft#page=Project/tolerance characterization of BBa_K398108] has been described on the TU Delft iGEM Team 2010 wiki. | ||
===Results=== | ===Results=== | ||
Up to 0.2 M NaCl effects of salt stress are equally observed for both the negative control and for cells with our biobrick ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K398108 BBa_K398108]). At higher concentrations a significant improvement of growth rate in comparison to the control background ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K398027 BBa_K398027]) is seen. In conclusion While at low salt concentrations no phenotype is observed, the resistance to high salt concentrations is significantly improved (up to 35%). The observed behavior can be explained by the vast amount of effects resulting from salt stress. It is possible that our BioBrick assists to reduce one of the inhibiting effects ,leading to a benefit at higher salt stress. | Up to 0.2 M NaCl effects of salt stress are equally observed for both the negative control and for cells with our biobrick ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K398108 BBa_K398108]). At higher concentrations a significant improvement of growth rate in comparison to the control background ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K398027 BBa_K398027]) is seen. In conclusion While at low salt concentrations no phenotype is observed, the resistance to high salt concentrations is significantly improved (up to 35%). The observed behavior can be explained by the vast amount of effects resulting from salt stress. It is possible that our BioBrick assists to reduce one of the inhibiting effects ,leading to a benefit at higher salt stress. | ||
| + | |||
| + | ===Further Characterization [iGEM Tuebingen 2019]=== | ||
| + | <html> | ||
| + | <b>BBa_K398108 salt tolerance cluster does not confer to an increased tolerance to potassium chloride.</b> | ||
| + | |||
| + | <p>We inoculated overnight cultures of <i>E. coli</i> DH5alpha wild type and salt tolerant DH5alpha in LB medium containing 0M to 0.8M KCl. The data was gained in triplicates and measured in OD600 with Implen’s Nanospectrometer every 45 minutes (in accordance with the previous characterization for NaCl). For the analysis OD600 values were transformed to logarithmic values and subsequently nonlinearly fitted to a growth curve using GraphPad Prism8.0.</p> | ||
| + | |||
| + | <p><img src="https://2019.igem.org/wiki/images/e/e2/T--Tuebingen--BBa_K398108.png" alt="Results" style="float:right; width:600px">The graph shows the growth rate [h-1] and [%]-growth of salt tolerant <i>E. coli</i> DH5alpha and wild type (wt) DH5alpha. The growth rate was calculated by using the proposed doubling time of the fit. [%]-growth was calculated by the normalisation of the growth rate of the salt tolerant DH5alpha against the wild type DH5alpha. The results came from triplicates for each concentration of potassium chloride.</p> | ||
| + | |||
| + | <p>The graph shows that wild type DH5alpha has approx. 20% higher growth rates in all compared concentrations, except 0.8M KCl, where the reduction of DH5alpha wild type is stronger.</p> | ||
| + | |||
| + | <p>Additionally, we expressed growth rates in [%]-growth by a normalisation of the growth rates of DH5alpha with salt tolerance against the wild type growth rates. When looking at [%]-growth, one may observe a slight increase in [%]-growth of the salt tolerant DH5alpha upon addition of KCl. This may indicate that the salt tolerant bacteria are less inhibited in their growth, however these findings were not significant. To summarize, this suggests that the salt tolerance cluster does not confer to significant KCl tolerance in DH5alpha cells.</p> | ||
| + | </html> | ||
===References=== | ===References=== | ||
| Line 21: | Line 34: | ||
#'''Y. Hase, S. Yokoyama, A. Muto, et al.''' , Removal of a ribosome small subunit-dependent GTPase confers salt resistance on Escherichia coli cells. ''RNA Society'', 15:1766-1774 ('''2009''') | #'''Y. Hase, S. Yokoyama, A. Muto, et al.''' , Removal of a ribosome small subunit-dependent GTPase confers salt resistance on Escherichia coli cells. ''RNA Society'', 15:1766-1774 ('''2009''') | ||
| + | ===More Information (added by WVHS_San_Diego 2021)=== | ||
| + | |||
| + | <html><body> | ||
| + | |||
| + | <p><b> Summary: </b> | ||
| + | Provided background information of the bbc1 protein from literature regarding its first discovery and tolerance against salt-stress, to help with the understanding of its function. | ||
| + | </b> </p> | ||
| + | |||
| + | |||
| + | <p> <b> Documentation: </b> </p> | ||
| + | <p> In its first discovery, the bbc 1 gene was first taken interest in after it was more highly expressed in benign compared to carcinoma breast tumors, as determined by in situ hybridization to tissue sections (Adams et al). Eventually, the eukaryotic bbc1 protein was found to be expressed in a variety of organisms, from Drosophila to Arabidopsis Thaliana (Helps et al, Bertauche et al). | ||
| + | Specifically, this part’s protein was chosen from algal Chlamydomonas sp. W80, which increases the salt tolerance of E.coli. This was derived from the experiment conducted by Tanaka et al, which both confirmed the algal bbc1 gene in the E.coli as well as examined the stress tolerance (to both temperature and salinity conditions) of the E. coli cells. Since both salt-stress and colder temperature stress cause intracellular dehydration and inflict damage to the proteins and membranes, it was crucial that the results yielded an increased tolerance against the salt and freezing stress for the E. coli cells expressing the bbc1 gene. Both its hydrophilic property of the protein or perhaps its transcription-activating motif (bbc1 activates transcriptional activity corresponding to stress response genes in E. coli), were possible explanations for the function of the bbc1 protein. However, based on the insignificant difference between the metabolic changes of the control and cells expressing bbc1, Tanaka et al were able to determine that metabolic changes were not correlated with the acquisition of the tolerance developed by E. coli (Tanaka et al). | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <p><b> References </b> | ||
| + | Adams, S.m., Helps, N.r., Sharp, M., Brammar, W.j., Walker, R.a., & Varley, J.m. (1992). Isolation and characterization of a novel gene with differential expression in benign and malignant human breast tumours. Human Molecular Genetics, 1(2), 91-96. https://doi.org/10.1093/hmg/1.2.91 | ||
| + | Bertauche, N., Leung, J., & Giraudat, J. (1994). Conservation of the human breast basic conserved 1 gene in the plant kingdom: Characterization ofacDNA clone from arabidopsis thaliana. Gene, 141(2), 211-214. https://doi.org/10.1016/0378-1119(94)90573-8 | ||
| + | Helps, N.r., Adams, S.m., Brammar, W.j., & Varley, J.m. (1995). The drosophila melanogaster homologue of the human bbc1 gene is highly expressed during embryogenesis. Gene, 162(2), 245-248. https://doi.org/10.1016/0378-1119(95)00356-b | ||
| + | Tanaka, S., Ikeda, K., & Miyasaka, H. (2001). Enhanced tolerance against salt-stress and freezing-stress of escherichia coli cells expressing algal bbc1 gene. Current Microbiology, 42(3), 173-177. https://doi.org/10.1007/s002840010199 | ||
| + | </b> </p> | ||
| + | |||
| + | </body></html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ===Characterization of KCl tolerance=== | |
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 06:06, 21 October 2021
Salt tolerance cluster
BBa_K398108 enables E.coli to tolerate higher Sodium Chloride concentrations, the growth rate increase compared to a negative control was 35% for a 0.5M Sodium Chloride concentration.
Introduction
Salts are essential for life, but not all life requires the same amount or the same type of salts. But because of these varying conditions the salt stress differs from environment to environment. It is difficult for a species to adapt to different salt stress levels. To compensate for the cells innate vulnerability to higher salt concentrations we chose a protein from Chlamydomonas sp. W80 that increases the salt tolerance of E.coli. This protein is called bbc1.The exact mechanism of the increased salt tolerance is as of yet unknown. But since bbc1 shows a high homology to RBS binding proteins, it is theorized that it may prevent/stabilize the folding structure of the ribosome under high salt stress conditions.
Characterization
The salt tolerance induces by this BioBrick was tested by using NaCl in LB and M9 (containing 40 mM casaminoacids) media from 0 to 1M salt concentrations, it was compared to another of the TU Delft 2010 BioBricks which had the same antibiotic resistance, and same expression level (but of another protein which did not cause increased salt tolerance). abc The [http://2010.igem.org/Team:TU_Delft#page=Project/tolerance characterization of BBa_K398108] has been described on the TU Delft iGEM Team 2010 wiki.
Results
Up to 0.2 M NaCl effects of salt stress are equally observed for both the negative control and for cells with our biobrick (BBa_K398108). At higher concentrations a significant improvement of growth rate in comparison to the control background (BBa_K398027) is seen. In conclusion While at low salt concentrations no phenotype is observed, the resistance to high salt concentrations is significantly improved (up to 35%). The observed behavior can be explained by the vast amount of effects resulting from salt stress. It is possible that our BioBrick assists to reduce one of the inhibiting effects ,leading to a benefit at higher salt stress.
Further Characterization [iGEM Tuebingen 2019]
BBa_K398108 salt tolerance cluster does not confer to an increased tolerance to potassium chloride.
We inoculated overnight cultures of E. coli DH5alpha wild type and salt tolerant DH5alpha in LB medium containing 0M to 0.8M KCl. The data was gained in triplicates and measured in OD600 with Implen’s Nanospectrometer every 45 minutes (in accordance with the previous characterization for NaCl). For the analysis OD600 values were transformed to logarithmic values and subsequently nonlinearly fitted to a growth curve using GraphPad Prism8.0.
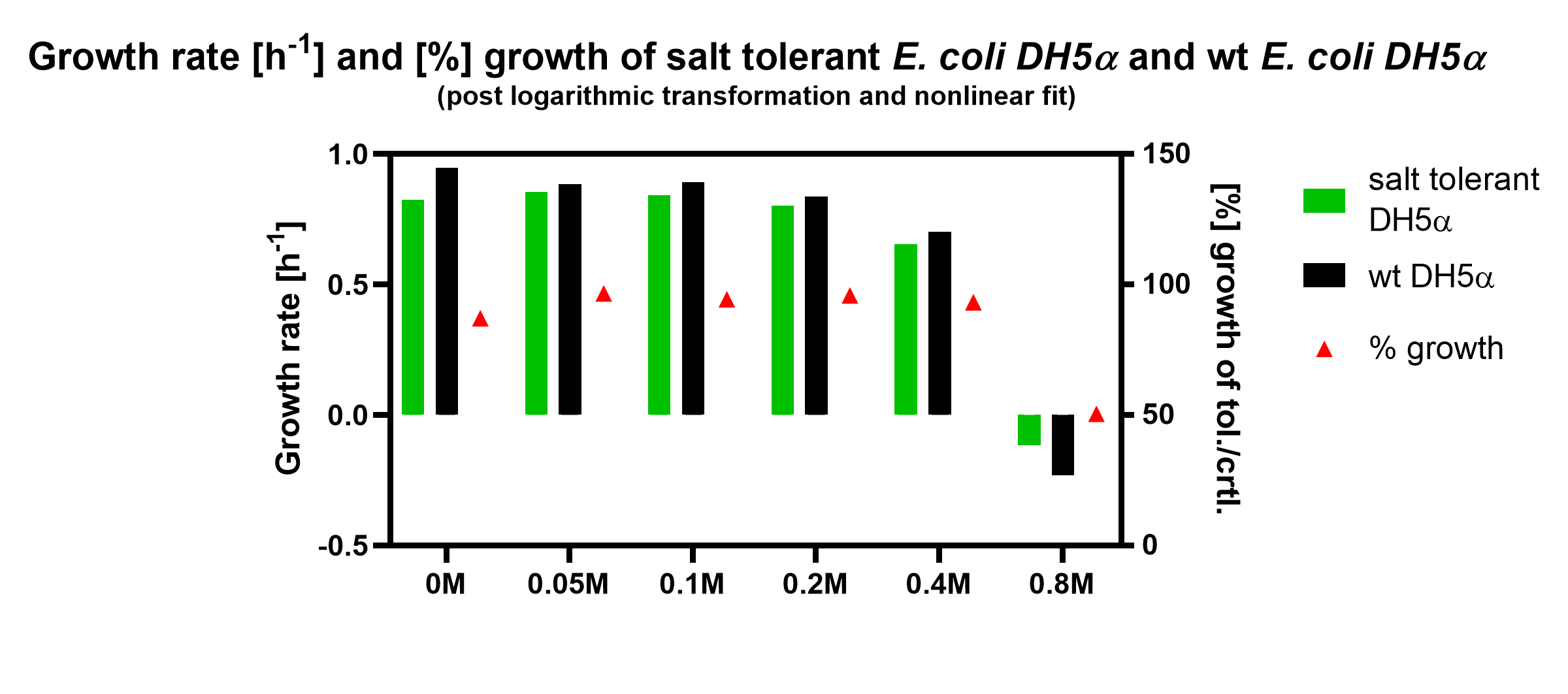 The graph shows the growth rate [h-1] and [%]-growth of salt tolerant E. coli DH5alpha and wild type (wt) DH5alpha. The growth rate was calculated by using the proposed doubling time of the fit. [%]-growth was calculated by the normalisation of the growth rate of the salt tolerant DH5alpha against the wild type DH5alpha. The results came from triplicates for each concentration of potassium chloride.
The graph shows the growth rate [h-1] and [%]-growth of salt tolerant E. coli DH5alpha and wild type (wt) DH5alpha. The growth rate was calculated by using the proposed doubling time of the fit. [%]-growth was calculated by the normalisation of the growth rate of the salt tolerant DH5alpha against the wild type DH5alpha. The results came from triplicates for each concentration of potassium chloride.
The graph shows that wild type DH5alpha has approx. 20% higher growth rates in all compared concentrations, except 0.8M KCl, where the reduction of DH5alpha wild type is stronger.
Additionally, we expressed growth rates in [%]-growth by a normalisation of the growth rates of DH5alpha with salt tolerance against the wild type growth rates. When looking at [%]-growth, one may observe a slight increase in [%]-growth of the salt tolerant DH5alpha upon addition of KCl. This may indicate that the salt tolerant bacteria are less inhibited in their growth, however these findings were not significant. To summarize, this suggests that the salt tolerance cluster does not confer to significant KCl tolerance in DH5alpha cells.
References
- S. Tanaka,K. Ikeda, H. Miyasaka, Enhanced Tolerance Against Salt-Stress and Freezing-Stress of Escherichia coli Cells Expressing Algal bbc1 Gene. Current Microbiology, 42:173-177 (2001)
- Y. Suda,T. Yoshikawa,Y. Okuda,M. Tsunemoto,S. Tanaka,K. Ikeda,H. Miyasaka,M. Watanabe,K. Sasaki,K. Harada,T. Bamba,K. Hirata, Isolation and characterization of a novel antistress gene from Chlamydomonas sp. W80. Journal of Bioscience and Bioengineering, 107(4) 352-354 (2009)
- Y. Hase, S. Yokoyama, A. Muto, et al. , Removal of a ribosome small subunit-dependent GTPase confers salt resistance on Escherichia coli cells. RNA Society, 15:1766-1774 (2009)
More Information (added by WVHS_San_Diego 2021)
Summary: Provided background information of the bbc1 protein from literature regarding its first discovery and tolerance against salt-stress, to help with the understanding of its function.
Documentation:
In its first discovery, the bbc 1 gene was first taken interest in after it was more highly expressed in benign compared to carcinoma breast tumors, as determined by in situ hybridization to tissue sections (Adams et al). Eventually, the eukaryotic bbc1 protein was found to be expressed in a variety of organisms, from Drosophila to Arabidopsis Thaliana (Helps et al, Bertauche et al). Specifically, this part’s protein was chosen from algal Chlamydomonas sp. W80, which increases the salt tolerance of E.coli. This was derived from the experiment conducted by Tanaka et al, which both confirmed the algal bbc1 gene in the E.coli as well as examined the stress tolerance (to both temperature and salinity conditions) of the E. coli cells. Since both salt-stress and colder temperature stress cause intracellular dehydration and inflict damage to the proteins and membranes, it was crucial that the results yielded an increased tolerance against the salt and freezing stress for the E. coli cells expressing the bbc1 gene. Both its hydrophilic property of the protein or perhaps its transcription-activating motif (bbc1 activates transcriptional activity corresponding to stress response genes in E. coli), were possible explanations for the function of the bbc1 protein. However, based on the insignificant difference between the metabolic changes of the control and cells expressing bbc1, Tanaka et al were able to determine that metabolic changes were not correlated with the acquisition of the tolerance developed by E. coli (Tanaka et al).
References Adams, S.m., Helps, N.r., Sharp, M., Brammar, W.j., Walker, R.a., & Varley, J.m. (1992). Isolation and characterization of a novel gene with differential expression in benign and malignant human breast tumours. Human Molecular Genetics, 1(2), 91-96. https://doi.org/10.1093/hmg/1.2.91 Bertauche, N., Leung, J., & Giraudat, J. (1994). Conservation of the human breast basic conserved 1 gene in the plant kingdom: Characterization ofacDNA clone from arabidopsis thaliana. Gene, 141(2), 211-214. https://doi.org/10.1016/0378-1119(94)90573-8 Helps, N.r., Adams, S.m., Brammar, W.j., & Varley, J.m. (1995). The drosophila melanogaster homologue of the human bbc1 gene is highly expressed during embryogenesis. Gene, 162(2), 245-248. https://doi.org/10.1016/0378-1119(95)00356-b Tanaka, S., Ikeda, K., & Miyasaka, H. (2001). Enhanced tolerance against salt-stress and freezing-stress of escherichia coli cells expressing algal bbc1 gene. Current Microbiology, 42(3), 173-177. https://doi.org/10.1007/s002840010199
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 312

