Difference between revisions of "Part:BBa K823037"
(→Methods) |
|||
| (One intermediate revision by one other user not shown) | |||
| Line 29: | Line 29: | ||
| − | Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''BacillusB'''''io'''B'''rick'''B'''ox]. | + | Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''BacillusB'''''io'''B'''rick'''B'''ox]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The ''Bacillus'' BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with ''Bacillus subtilis''] by Radeck ''et al.''. |
===Evaluation=== | ===Evaluation=== | ||
| Line 73: | Line 73: | ||
<br> | <br> | ||
| + | ==Improvement== | ||
| + | ===Improvement by IISc-Bangalore 2021=== | ||
| + | This part was codon-optimized for expression in ''E. coli''. We attached this 10xHis Tag to our SpyCatcher002-dCBD and OpdA-SpyTag002 constructs, and purified them via Ni-NTA affinity column chromatography. We also carried out Western blots for these fusion proteins using anti-His primary antibodies which could reliably detect this tag. See [https://parts.igem.org/Part:BBa_K3765003 BBa_K3765003] for more details. | ||
| + | |||
| + | ==Link== | ||
| + | |||
| + | 【1】https://parts.igem.org/Part:BBa_K3765003 | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 78: | Line 85: | ||
<!-- --> | <!-- --> | ||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K823037 SequenceAndFeatures</partinfo> | <partinfo>BBa_K823037 SequenceAndFeatures</partinfo> | ||
Latest revision as of 05:59, 21 October 2021
10x His-tag (Freiburg standard+RBS)
10xHis-tag with RBS in Freiburg standard.
Find out more about the design of our prefix with ribosome binding site.
prefix:GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix:ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
The His-tag is a metal chelating peptide ([http://www.nature.com/nbt/journal/v6/n11/full/nbt1188-1321.html Hochuli, E.; Bannwarth, W.; Döbeli, H.; Gentz, R.; Stüber, D. (1988)]) consisting of at least 6 histidins. It can therefore be used for protein purification by metal 2+ (mostly nickel or cobalt) containing columns. There are also antibodies against this tag, or nickel/cobalt containing fluorescent probes can be used for detection. Also a immobilization is possible in nickel/cobalt coated plastikware. The aminoacid sequence is:HHHHHHHHHH
This is a part created by the LMU-Munich 2012 team. We added five tags to the registry, all in the Freiburg standard for N-and C-terminal fusions:
- His - tag
Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks BacillusBioBrickBox]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
Evaluation
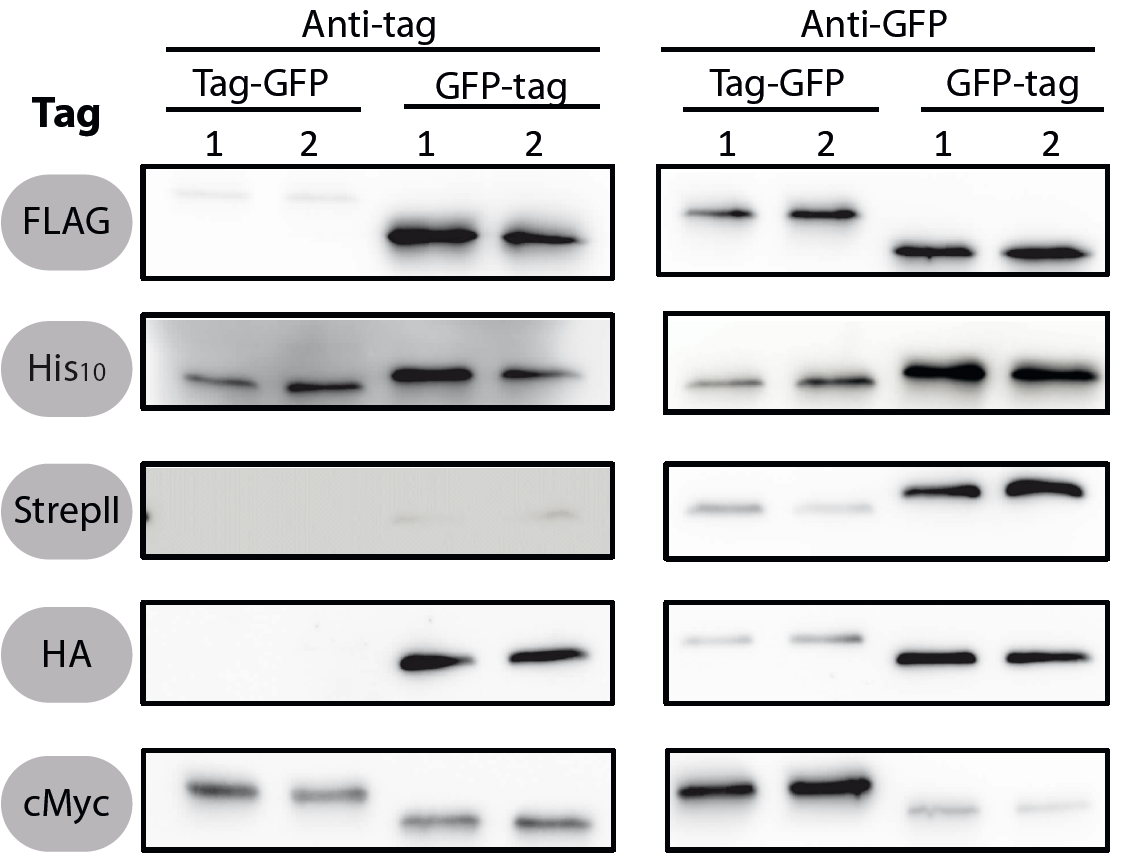
All 5 epitope tags were fused C- and N-terminally to GFP using the NgoMIV and AgeI restriction sites. These constructs were expressed in Bacillus subtils using pSBBs0K-Pspac. This vector did not need to be induced by IPTG due to a premature stop codon in the lacI gene.
|
Methods
To verify the functionality of the epitope tags, Western blot analyses of the strains TMB1920-TMB1929 were performed. LB medium (15 ml) was inoculated 1:100 from overnight culture and grown at 37°C and 200 rpm to OD600 ~ 0.5. Of this, 10 ml were harvested by centrifugation (8000 × g, 5 min) and the pellets stored at -20°C. Pellets were resuspended in 1 ml disruption buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl) and lysed by sonication. Samples (12 μl of lysate) were loaded per lane on two 12.5% SDS-polyacrylamide gels and SDS-PAGE was performed according standard procedure [60]. One gel was stained with colloidal coomassie, the other one was used for protein transfer to a PVDF membrane (Merck Millipore, Billerica, MA, USA) by submerged blotting procedure (Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA)). After protein transfer, the membranes were treated with the following antibodies and conditions. Detailed protocols can be found [http://www.jbioleng.org/content/7/1/29/suppl/S3 here].
GFP
Probing with primary antibodies takes place with rabbit anti-GFP antibodies (1:3000, Epitomics, No. 1533). Horseradish-peroxidase (HRP)-conjugated anti-rabbit antibodies (1:2000, Promega, W401B) were used as secondary antibody. Hybridization of both antibodies was carried out in Blotto-buffer (2.5% (w/v) skim milk powder, 1 × TBS (50 mM Tris–HCl pH 7.6, 0.15 M NaCl)).
His10
Mouse-anti-Penta-His (1:2000, Qiagen, Penta-His, No. 34660) in 1 × TBS, 5% bovine serum albumin (BSA, Fraction V; Carl ROTH) and anti-mouse-HRP (1:2000, Promega, W402B1) in 1 × TBS, 10 % (w/v) skim milk powder were used.
Chemiluminescence signals were detected after addition of the HRP-substrate Ace Glow (Peqlab, Erlangen, Germany) using a FusionTM imaging system (Peqlab).
Improvement
Improvement by IISc-Bangalore 2021
This part was codon-optimized for expression in E. coli. We attached this 10xHis Tag to our SpyCatcher002-dCBD and OpdA-SpyTag002 constructs, and purified them via Ni-NTA affinity column chromatography. We also carried out Western blots for these fusion proteins using anti-His primary antibodies which could reliably detect this tag. See BBa_K3765003 for more details.
Link
【1】https://parts.igem.org/Part:BBa_K3765003
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]