Difference between revisions of "Part:BBa K3247005"
HanZhenhao (Talk | contribs) |
|||
| (3 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| + | __NOTOC__ | ||
| + | <partinfo>BBa_K3247005 short</partinfo> | ||
| + | RNA thermometers are a form of translational regulation. This RNA thermometer was designed to have a high fold change between 25°C and 30°C, but ended up having a high fold change between 25 °C and 37 °C. It is an improvement upon the part BBa_K11502. | ||
| + | |||
| + | ===Design Notes=== | ||
| + | The stem-loop structure of the thermometer was created by taking the complement of the ribosome binding site (RBS) and adding nucleotides to either side. The additional bases were mutated; the resulting altered sequence is known as the variable region. Mutating the variable region keeps the RBS intact while modifying the RNA thermometer secondary structure to change the melting temperature. There is no internal BsaI cut site, since it would render the thermometer incompatible with our assembly method. The part is TypeIIS compatible. Figure 1a-c showing the RNA thermometer structures were designed using NUPACK software. | ||
| + | |||
| + | <html> | ||
| + | <img src="https://2019.igem.org/wiki/images/9/98/T--Rice--0625.png" width = 60%> | ||
| + | <p><b>Figure 1a.</b> Predicted RNA thermometer structure at 25 °C.</p> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/27/T--Rice--0630.png" width = 60%> | ||
| + | <p><b>Figure 1b.</b> Predicted RNA thermometer structure at 30 °C.</p> | ||
| + | <img src="https://2019.igem.org/wiki/images/0/03/T--Rice--0637.png" width = 60%> | ||
| + | <p><b>Figure 1c.</b> Predicted RNA thermometer structure at 37 °C.</p> | ||
| + | </html> | ||
| + | |||
| + | ===Characterization=== | ||
| + | <html> | ||
| + | <img src="https://2019.igem.org/wiki/images/0/0e/T--Rice--Nochill6.png" width = 60%> | ||
| + | <p><b>Figure 2.</b> Fluorescence intensity at 25 °C, 30 °C, and 37 °C.</p> | ||
| + | </html> | ||
| + | |||
| + | ==<h3><b>Contribution From HZAU-China 2021</b></h3>== | ||
| + | === Results and analysis=== | ||
| + | <p> | ||
| + | To verify that the RNA thermometer could work, we constructed pUC57-J23110-RNA thermometer-neGFP (BBa_K3733012) plasmid and transformed it into <i>E.coli</i> DH5α. We cultured both experimental group and control group at both 37 ℃ and 28 ℃ for 12 hours and detected RFU / OD<sub>600</sub> by a microplate reader. As the result shown in <b>Figure 1</b>, RFU / OD<sub>600</sub> data of the media at 37 ℃ are obviously higher than ones at 28 ℃, which could explain the RNA thermometer is valid. | ||
| + | </p> | ||
| + | |||
| + | <html> | ||
| + | <head> | ||
| + | <meta charset="utf-8"> | ||
| + | <title>无标题文档</title> | ||
| + | </head> | ||
| + | <body> | ||
| + | <center><img src="https://static.igem.org/mediawiki/parts/9/90/T--HZAU-China-T-ind-comp-1.png" style="width:500px;height:360px"></center> | ||
| + | <center><b>Figure 1. </b> RFU / OD<sub>600</sub> data of the media at 37 ℃ and 28 ℃(heat-inducible RNA thermosensors). </center> | ||
| + | <br> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | <partinfo>BBa_K3247005 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ===Source=== | ||
| + | |||
| + | The thermometers were designed de novo using NUPACK and VSAlgorithm | ||
| + | |||
| + | |||
| + | ===References=== | ||
| + | J. N. Zadeh, C. D. Steenberg, J. S. Bois, B. R. Wolfe, M. B. Pierce, A. R. Khan, R. M. Dirks, N. A. Pierce. NUPACK: analysis and design of nucleic acid systems. J Comput Chem, 32:170–173, 2011. (pdf) | ||
Latest revision as of 18:48, 20 October 2021
RNA Thermometer NoChill-06
RNA thermometers are a form of translational regulation. This RNA thermometer was designed to have a high fold change between 25°C and 30°C, but ended up having a high fold change between 25 °C and 37 °C. It is an improvement upon the part BBa_K11502.
Design Notes
The stem-loop structure of the thermometer was created by taking the complement of the ribosome binding site (RBS) and adding nucleotides to either side. The additional bases were mutated; the resulting altered sequence is known as the variable region. Mutating the variable region keeps the RBS intact while modifying the RNA thermometer secondary structure to change the melting temperature. There is no internal BsaI cut site, since it would render the thermometer incompatible with our assembly method. The part is TypeIIS compatible. Figure 1a-c showing the RNA thermometer structures were designed using NUPACK software.

Figure 1a. Predicted RNA thermometer structure at 25 °C.
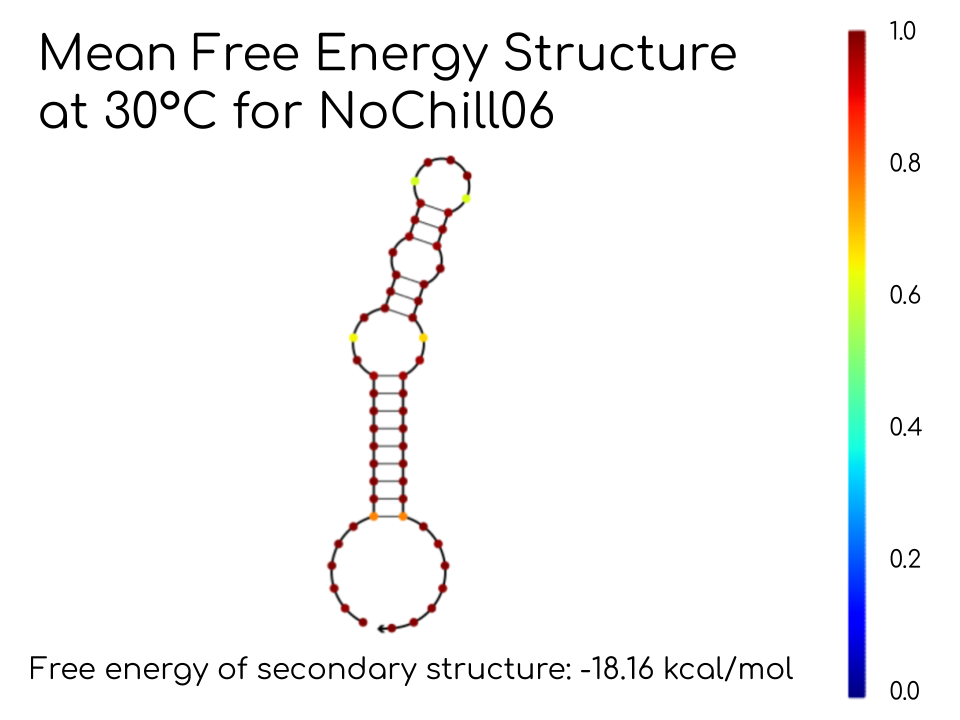
Figure 1b. Predicted RNA thermometer structure at 30 °C.

Figure 1c. Predicted RNA thermometer structure at 37 °C.
Characterization

Figure 2. Fluorescence intensity at 25 °C, 30 °C, and 37 °C.
Contribution From HZAU-China 2021
Results and analysis
To verify that the RNA thermometer could work, we constructed pUC57-J23110-RNA thermometer-neGFP (BBa_K3733012) plasmid and transformed it into E.coli DH5α. We cultured both experimental group and control group at both 37 ℃ and 28 ℃ for 12 hours and detected RFU / OD600 by a microplate reader. As the result shown in Figure 1, RFU / OD600 data of the media at 37 ℃ are obviously higher than ones at 28 ℃, which could explain the RNA thermometer is valid.

- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Source
The thermometers were designed de novo using NUPACK and VSAlgorithm
References
J. N. Zadeh, C. D. Steenberg, J. S. Bois, B. R. Wolfe, M. B. Pierce, A. R. Khan, R. M. Dirks, N. A. Pierce. NUPACK: analysis and design of nucleic acid systems. J Comput Chem, 32:170–173, 2011. (pdf)
