Difference between revisions of "Part:BBa K3796219"
| Line 18: | Line 18: | ||
====1.Identification==== | ====1.Identification==== | ||
<p>From the chemically synthesized sequence, we obtained the fragment from the plasmid by PCR and recovered the gel.</p> | <p>From the chemically synthesized sequence, we obtained the fragment from the plasmid by PCR and recovered the gel.</p> | ||
| − | [[image:T--CAU China--Parts04 gel.png|400px]] | + | [[image:T--CAU China--Parts04 gel.png|400px|thumb|center|]] |
| − | <p>Fig.1 P0864 electrophoresis band. Lane 1 to 4.</p> | + | <p style="text-align: center;"><b>Fig.1 P0864 electrophoresis band. Lane 1 to 4.</b></p> |
<br> | <br> | ||
====2.Strength identification==== | ====2.Strength identification==== | ||
<p>In order to detect the activity of the new promoter P0864 in <i>Corynebacterium glutamicum</i> and <i>Escherichia coli</i>, we constructed the related circuit in pXMJ19 plasmid. The expression of P0864 was characterized by fluorescence size.</p> | <p>In order to detect the activity of the new promoter P0864 in <i>Corynebacterium glutamicum</i> and <i>Escherichia coli</i>, we constructed the related circuit in pXMJ19 plasmid. The expression of P0864 was characterized by fluorescence size.</p> | ||
| − | [[image:T--CAU China--Part01 pmc.svg| | + | [[image:T--CAU China--Part01 pmc.svg|600px|thumb|center|]] |
| − | <p>Fig.2 Experimental group gene circuit</p> | + | <p style="text-align: center;"><b>Fig.2 Experimental group gene circuit</b></p> |
<p>Since pXMJ19 itself contains an inducible promoter PTAC (BBa_m31370), and the inducible promoter may be leaked, it is necessary to construct a gene line without P0864 as a control.</p> | <p>Since pXMJ19 itself contains an inducible promoter PTAC (BBa_m31370), and the inducible promoter may be leaked, it is necessary to construct a gene line without P0864 as a control.</p> | ||
| − | [[image:T--CAU China--Part02 mc.svg| | + | [[image:T--CAU China--Part02 mc.svg|600px|thumb|center|]] |
| − | <p>Fig.3 Control group gene circuit</p> | + | <p style="text-align: center;"><b>Fig.3 Control group gene circuit</b></p> |
<br> | <br> | ||
<p>The transformed strains were cultured on LBG chloramphenicol resistant solid medium for 24 hours. In <i>E.coli</i>, two gene circuits showed different fluorescence. It has different performance under the irradiation of different light sources. The fluorescence of circuit containing P0864 is significantly brighter than that of another circuit. Under the irradiation of natural light, the colonies showed purplish red.</p> | <p>The transformed strains were cultured on LBG chloramphenicol resistant solid medium for 24 hours. In <i>E.coli</i>, two gene circuits showed different fluorescence. It has different performance under the irradiation of different light sources. The fluorescence of circuit containing P0864 is significantly brighter than that of another circuit. Under the irradiation of natural light, the colonies showed purplish red.</p> | ||
| − | [[image:T--CAU China--Part03 expr&ck.png|400px]] | + | [[image:T--CAU China--Part03 expr&ck.png|400px|thumb|center|]] |
| − | <p>Fig.4 The difference of fluorescent protein expressed by positive <i>Escherichia coli</i> (including experimental group and control group). a. Image under excitation light.(Left: Experimental group, right: control group) b. Images in natural light.(Left: Experimental group, right: control group)</p> | + | <p style="text-align: center;"><b>Fig.4 The difference of fluorescent protein expressed by positive <i>Escherichia coli</i> (including experimental group and control group). a. Image under excitation light.(Left: Experimental group, right: control group) b. Images in natural light.(Left: Experimental group, right: control group)</b></p> |
<br> | <br> | ||
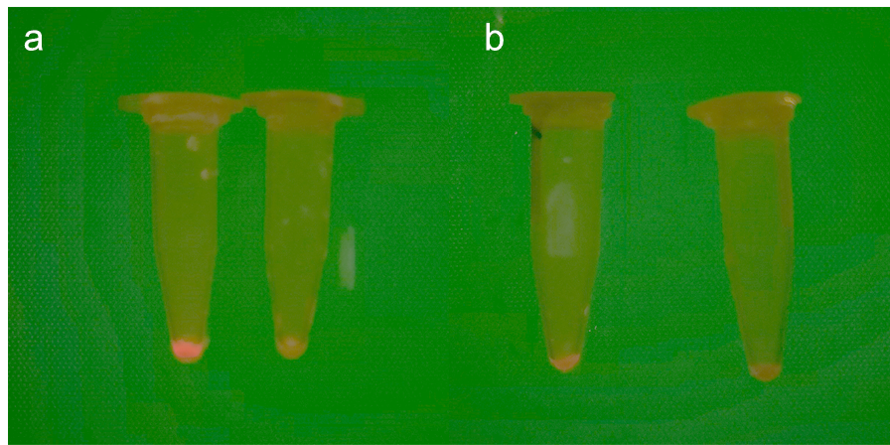
<p><i>Corynebacterium glutamicum</i> and <i>Escherichia coli</i> were cultured in LBG chloramphenicol liquid medium at 35 ℃ for 26h. The bacteria were collected by centrifugation and observed under light. Then the bacteria were resuspended, the fluorescence relative value of the bacterial solution was measured by fluorescence spectrophotometer, and its OD600 was measured.</p> | <p><i>Corynebacterium glutamicum</i> and <i>Escherichia coli</i> were cultured in LBG chloramphenicol liquid medium at 35 ℃ for 26h. The bacteria were collected by centrifugation and observed under light. Then the bacteria were resuspended, the fluorescence relative value of the bacterial solution was measured by fluorescence spectrophotometer, and its OD600 was measured.</p> | ||
| − | [[image:T--CAU China--Parts tube.png|400px]] | + | [[image:T--CAU China--Parts tube.png|400px|thumb|center|]] |
| − | <p>Fig.5 Difference of fluorescent proteins expressed by positive <i>Escherichia coli</i> and <i>Corynebacterium glutamicum</i> (including experimental group and control group). a. Images of <i>Corynebacterium glutamicum</i> after centrifugation, including experimental group and control group. b. Images of <i>E.coli</i> after centrifugation, including experimental group and control group.</p> | + | <p style="text-align: center;"><b>Fig.5 Difference of fluorescent proteins expressed by positive <i>Escherichia coli</i> and <i>Corynebacterium glutamicum</i> (including experimental group and control group). a. Images of <i>Corynebacterium glutamicum</i> after centrifugation, including experimental group and control group. b. Images of <i>E.coli</i> after centrifugation, including experimental group and control group.</b></p> |
<br> | <br> | ||
| − | [[image:T--CAU China--Parts data.png|400px]] | + | [[image:T--CAU China--Parts data.png|400px|thumb|center|]] |
| − | <p>Fig.6 After <i>E.coli</i> and <i>C.glutamicum</i> were cultured at 35 ℃ for 27 hours, the ratio of measured fluorescence data to OD600 data.</p> | + | <p style="text-align: center;"><b>Fig.6 After <i>E.coli</i> and <i>C.glutamicum</i> were cultured at 35 ℃ for 27 hours, the ratio of measured fluorescence data to OD600 data.</b></p> |
<br> | <br> | ||
<p>T-test was performed on the three groups of data.</p> | <p>T-test was performed on the three groups of data.</p> | ||
| − | <p>Table 1 T-test results of two species. ***: p-value<0.001, **: p-value<0.01</p> | + | <p style="text-align: center;"><b>Table.1 T-test results of two species. ***: p-value<0.001, **: p-value<0.01</b></p> |
| − | [[image:T--CAU China--Parts ttest.png|400px]] | + | [[image:T--CAU China--Parts ttest.png|400px|thumb|center|]] |
<p>The mean value of experimental group was significantly different from that of control group. The data showed that P0864 could be expressed in <i>E.coli</i>, and the expression amount was significantly different from that in the control group. As a constitutive promoter, it can be used as a standardized element.</p> | <p>The mean value of experimental group was significantly different from that of control group. The data showed that P0864 could be expressed in <i>E.coli</i>, and the expression amount was significantly different from that in the control group. As a constitutive promoter, it can be used as a standardized element.</p> | ||
Revision as of 16:43, 20 October 2021
P0864 strength measurement
This sequence was used for the intensity measurement of P0864. The mCherry protein was expressed with P0864, and then the fluorescence was measured.
Usage
P0864 was used to express capA capB capC genes in our project. The promoter is a strong constitutive expression promoter, which can be well applied to the constitutive expression of genes, whether in Escherichia coli or Corynebacterium glutamicum. When the sequence to be transcribed is installed downstream of p0864, the sequence will be transcribed with a strong intensity without the action of inducer.
Biology
P0864 is an endogenous constitutive expression promoter of Corynebacterium glutamicum, which can be recognized by bacterial sigma factor and start downstream gene transcription.
Characterization
1.Identification
From the chemically synthesized sequence, we obtained the fragment from the plasmid by PCR and recovered the gel.
Fig.1 P0864 electrophoresis band. Lane 1 to 4.
2.Strength identification
In order to detect the activity of the new promoter P0864 in Corynebacterium glutamicum and Escherichia coli, we constructed the related circuit in pXMJ19 plasmid. The expression of P0864 was characterized by fluorescence size.
Fig.2 Experimental group gene circuit
Since pXMJ19 itself contains an inducible promoter PTAC (BBa_m31370), and the inducible promoter may be leaked, it is necessary to construct a gene line without P0864 as a control.
Fig.3 Control group gene circuit
The transformed strains were cultured on LBG chloramphenicol resistant solid medium for 24 hours. In E.coli, two gene circuits showed different fluorescence. It has different performance under the irradiation of different light sources. The fluorescence of circuit containing P0864 is significantly brighter than that of another circuit. Under the irradiation of natural light, the colonies showed purplish red.
Fig.4 The difference of fluorescent protein expressed by positive Escherichia coli (including experimental group and control group). a. Image under excitation light.(Left: Experimental group, right: control group) b. Images in natural light.(Left: Experimental group, right: control group)
Corynebacterium glutamicum and Escherichia coli were cultured in LBG chloramphenicol liquid medium at 35 ℃ for 26h. The bacteria were collected by centrifugation and observed under light. Then the bacteria were resuspended, the fluorescence relative value of the bacterial solution was measured by fluorescence spectrophotometer, and its OD600 was measured.
Fig.5 Difference of fluorescent proteins expressed by positive Escherichia coli and Corynebacterium glutamicum (including experimental group and control group). a. Images of Corynebacterium glutamicum after centrifugation, including experimental group and control group. b. Images of E.coli after centrifugation, including experimental group and control group.
Fig.6 After E.coli and C.glutamicum were cultured at 35 ℃ for 27 hours, the ratio of measured fluorescence data to OD600 data.
T-test was performed on the three groups of data.
Table.1 T-test results of two species. ***: p-value<0.001, **: p-value<0.01
The mean value of experimental group was significantly different from that of control group. The data showed that P0864 could be expressed in E.coli, and the expression amount was significantly different from that in the control group. As a constitutive promoter, it can be used as a standardized element.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]