Difference between revisions of "Part:BBa K3887009"
| Line 4: | Line 4: | ||
| − | |||
===Description=== | ===Description=== | ||
| Line 24: | Line 23: | ||
Meanwhile, though we made a fusion enzyme to improve SttH expression solubility, part of the enzymes could not be dissolved after expression and remained in the precipitation. | Meanwhile, though we made a fusion enzyme to improve SttH expression solubility, part of the enzymes could not be dissolved after expression and remained in the precipitation. | ||
We also did a fermentation experiment on 6,6-dichloroindigo using Fre-L3-SttH. According to the result, our manufacture of indigo derivatives was a great success. | We also did a fermentation experiment on 6,6-dichloroindigo using Fre-L3-SttH. According to the result, our manufacture of indigo derivatives was a great success. | ||
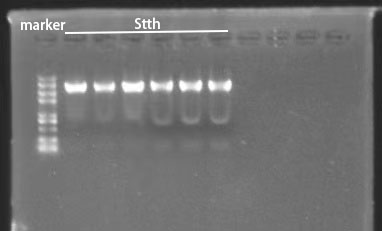
| − | [[File:T--BJU_China--Fig.4 SttH4.png| | + | [[File:T--BJU_China--Fig.4 SttH4.png|500px|thumb|center]] |
| − | <p class="figure-description"><b><center> Figure 4. Fermentation Results of Fre-L3-SttH | + | <p class="figure-description"><b><center> Figure 4. Fermentation Results of Fre-L3-SttH</center></b></p> |
Revision as of 03:42, 20 October 2021
Fre-L3-SttH
Description
SttH is an halogenases used during the conversion of indole or Tryptophan. It exhibits regiospecificitiy for both bromination and chloroindole at C6. However, when SttH overexpressed in vitro, most of the enzyme were exist in the precipitation for its low solubility. To improve this problem, we add Fre to increase its solubility with a rigid linker L3. Below is the gene circuit we used in our project.
Experiments and Results
We constructed halogenase plasmids using Gibson Assembly and verified by colony PCR and gene sequencing . According to the results in Figure 2, we can roughly identify the Fre-L3-SttH gene comparing with DNA Marker. And then the PCR product fragments were sent for sequencing and the results were also consistent with our target fragments.
Next, we want to test the expression of Fre-L3-SttH with IPTG induction. For better expression of enzymes, we optimize the induction condition of gene expression: three different temperatures (18℃, 30℃, 37℃) and inducer concentrations (0.1mM, 1mM, 5mM) were set for test. Additionally, since the 6-halogenase Fre-L3-SttH is a fusion enzyme (including core enzyme SttH,co-enzyme Fre and rigid linker L3) with higher solubility, we disrupted collected cells and tested both pellet and supernatant after induction (Figure 4).
From the results in Figure 4, we can see that Fre-L3-SttH was successfully expressed (arrow points). SttH induced by IPTG could express well at three different temperatures, and as the temperature increases, the expression level also gradually increases. There is no significant difference in the expression level of SttH at 37°C with 0.1, 1, and 5 mM IPTG, and the amount of protein is significantly more compared to the other two temperatures, so we can speculate that the optimal temperature for the expression of SttH is 37°C. Meanwhile, though we made a fusion enzyme to improve SttH expression solubility, part of the enzymes could not be dissolved after expression and remained in the precipitation. We also did a fermentation experiment on 6,6-dichloroindigo using Fre-L3-SttH. According to the result, our manufacture of indigo derivatives was a great success.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 1813
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 942
Illegal NgoMIV site found at 1200
Illegal NgoMIV site found at 1903
Illegal NgoMIV site found at 2169
Illegal AgeI site found at 1088
Illegal AgeI site found at 1429 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 301
Illegal SapI.rc site found at 421