Difference between revisions of "Part:BBa K1313004"
| (15 intermediate revisions by 3 users not shown) | |||
| Line 6: | Line 6: | ||
This gene can be also used as a selective gene for mammalian cells using gentamicin. | This gene can be also used as a selective gene for mammalian cells using gentamicin. | ||
| + | <html> | ||
| + | <h2>Contribution from other teams</h2> | ||
| + | <br> | ||
| + | <h3>Toulouse_INSA-UPS 2021's contribution</h3> | ||
| + | <br> | ||
| + | <h4>Characterization</h4> | ||
| + | <p> <a href="https://2021.igem.org/Team:Toulouse_INSA-UPS">Toulouse_INSA_UPS_2021</a>contributed to the characterization of this part. The part BBa_K1313004 for the <i>neoR</i> gene had not been characterized before in iGEM for yeast selection, but the team showed this year that it is functional for a transformant selection in <i>S. cerevisiae</i>. Check the part of the full construction <a href="https://parts.igem.org/Part:BBa_K3930002" class="pr-0" target="_blank">(BBa_K3930002)</a> for more result details.</p> | ||
| + | |||
| + | </html> | ||
| + | (<small>--[[User:ThomasG|ThomasG]] 16:53, 18 October 2021 (UTC+2)</small>) | ||
<h1> | <h1> | ||
Characterization iGEM 2019 | Characterization iGEM 2019 | ||
</h1> | </h1> | ||
| − | < | + | <h2> |
| − | By | + | By St Andrews 2019 |
| − | </ | + | </h2> |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<p> | <p> | ||
<h3 style="text-decoration:underline"> Aim | <h3 style="text-decoration:underline"> Aim | ||
</h3> | </h3> | ||
</p> | </p> | ||
| − | To characterize the part responsible for neomycin resistance (BBa_K1313004) and its response to kanamycin in DH5 alpha E. coli cells using OD measurements at 600 nm in the presence or absence of a plasmid containing the resistance sequence. | + | <p> |
| + | To characterize the part responsible for neomycin resistance (BBa_K1313004) and its response to kanamycin in DH5 alpha E. coli cells using OD measurements at 600 nm in the presence or absence of a plasmid containing the resistance sequence. | ||
| + | </p> | ||
| + | |||
<h3 style="text-decoration:underline"> Experimental design | <h3 style="text-decoration:underline"> Experimental design | ||
</h3> | </h3> | ||
| Line 29: | Line 36: | ||
Preparation: | Preparation: | ||
</h4> | </h4> | ||
| − | + | <p> | |
| + | PLBDE2 plasmid was used, however due to it being too big, a digestion was done followed by a ligation. The transformation had been concluded successful. However, the experiments with those cells were only successful if they were done with a double overnight culture. | ||
</p> | </p> | ||
| − | + | ||
<h4> | <h4> | ||
Experiment | Experiment | ||
</h4> | </h4> | ||
| + | <p> | ||
Bacteria with neomycin resistance and bacteria without such have been inoculated overnight. | Bacteria with neomycin resistance and bacteria without such have been inoculated overnight. | ||
8 vials were set up with different kanamycin concentrations ranging from 0 µg/µL to 200 µg/µL. In each one were added 100 µL of DH5alphas with neomycin resistance cassette. Another 8 vials with different kanamycin concentrations were prepared and 100 µL of non-resistant DH5 alphas were added in each vial. | 8 vials were set up with different kanamycin concentrations ranging from 0 µg/µL to 200 µg/µL. In each one were added 100 µL of DH5alphas with neomycin resistance cassette. Another 8 vials with different kanamycin concentrations were prepared and 100 µL of non-resistant DH5 alphas were added in each vial. | ||
| Line 50: | Line 59: | ||
<li> We can offer no explanation why the experiment was successful with only double overnight cultures </li> | <li> We can offer no explanation why the experiment was successful with only double overnight cultures </li> | ||
</ul> | </ul> | ||
| − | </ | + | </p> |
| + | |||
| + | <div align = "center"> | ||
| + | "https://2019.igem.org/wiki/images/1/1d/T--St_Andrews--neomycingraph.jpg" | ||
| + | <html> | ||
| + | <p style="text-align:center;"><font size=-2>Figure 1. Growth of neomycin resistance DH5alpha cells in different kanamycin concentrations | ||
| + | |||
| + | |||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 18:56, 19 October 2021
Neomycin Resistance
Neomycin resistance gene. This gene can be used for selection of cells with an efficient transformation. This gene codes neomycin phosphotransferase which breaks neomycin, kanamycin, and gentamicin molecules. If you use these gene as selectivity gene in bacteria, and your laboratory lacks neomycin, you can replace it for kanamycin by adding 35 mg/ml per liter of LB media. The best concentration of neomycin to use is 80 mg/ml per liter of LB media.
This gene can be also used as a selective gene for mammalian cells using gentamicin.
Contribution from other teams
Toulouse_INSA-UPS 2021's contribution
Characterization
Toulouse_INSA_UPS_2021contributed to the characterization of this part. The part BBa_K1313004 for the neoR gene had not been characterized before in iGEM for yeast selection, but the team showed this year that it is functional for a transformant selection in S. cerevisiae. Check the part of the full construction (BBa_K3930002) for more result details.
(--ThomasG 16:53, 18 October 2021 (UTC+2))Characterization iGEM 2019
By St Andrews 2019
Aim
To characterize the part responsible for neomycin resistance (BBa_K1313004) and its response to kanamycin in DH5 alpha E. coli cells using OD measurements at 600 nm in the presence or absence of a plasmid containing the resistance sequence.
Experimental design
Preparation:
PLBDE2 plasmid was used, however due to it being too big, a digestion was done followed by a ligation. The transformation had been concluded successful. However, the experiments with those cells were only successful if they were done with a double overnight culture.
Experiment
Bacteria with neomycin resistance and bacteria without such have been inoculated overnight. 8 vials were set up with different kanamycin concentrations ranging from 0 µg/µL to 200 µg/µL. In each one were added 100 µL of DH5alphas with neomycin resistance cassette. Another 8 vials with different kanamycin concentrations were prepared and 100 µL of non-resistant DH5 alphas were added in each vial. OD measurements at 600 nm were taken every 30 minutes to follow colony growth for 4 hours.
Experiment was repeated with higher kanamycin concentrations ranging from 0 µg/µL to 1000 µg/µL and the same observation as the previous experiment could be concluded.
Observations
- Cells with resistance grew overtime, and the vials containing high kanamycin concentrations produced slower cell growth
- Cells without resistance grew only in the vial with 0 µg/ µL kanamycin concentration
- We can offer no explanation why the experiment was successful with only double overnight cultures
"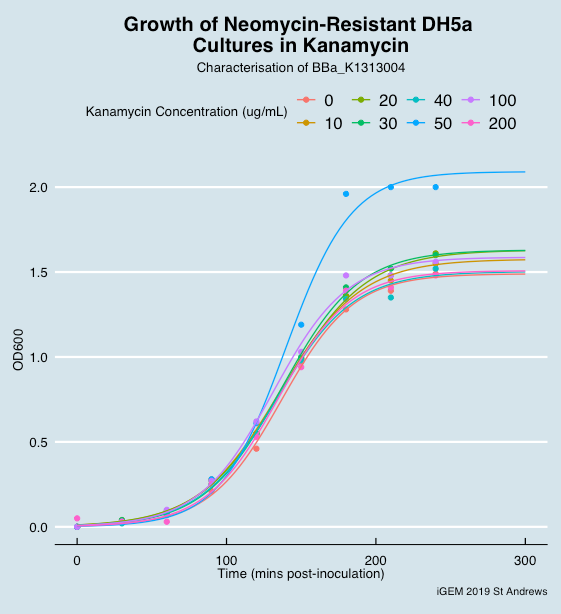 "
"
Figure 1. Growth of neomycin resistance DH5alpha cells in different kanamycin concentrations
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal PstI site found at 165
- 12INCOMPATIBLE WITH RFC[12]Illegal PstI site found at 165
- 21COMPATIBLE WITH RFC[21]
- 23INCOMPATIBLE WITH RFC[23]Illegal PstI site found at 165
- 25INCOMPATIBLE WITH RFC[25]Illegal PstI site found at 165
Illegal NgoMIV site found at 616 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 465
Illegal SapI.rc site found at 675
