Difference between revisions of "Part:BBa K3100017"
| (14 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3100017 short</partinfo> | <partinfo>BBa_K3100017 short</partinfo> | ||
| − | |||
| − | Usage and Biology: | + | <h2><strong>Usage and Biology:</strong></h2> |
GadB is an acid tolerant factor which play an important role in the acid tolerance of E.coli MG1655.The decarboxylation of glutamate consumes a proton, and therefore, micro‐organisms take advantage of this property to remove protons from the intracellular milieu under acidic conditions[1]. GadB is involved in the AR system, which is the glutamic acid-dependent acid resistance (GDAR) system, consisting of the homologous inducible glutamic acid decarboxylases GadA/GadB enzymes and the glutamate/γ-aminobutyric acid (GABA) antiporter GadC[2]. | GadB is an acid tolerant factor which play an important role in the acid tolerance of E.coli MG1655.The decarboxylation of glutamate consumes a proton, and therefore, micro‐organisms take advantage of this property to remove protons from the intracellular milieu under acidic conditions[1]. GadB is involved in the AR system, which is the glutamic acid-dependent acid resistance (GDAR) system, consisting of the homologous inducible glutamic acid decarboxylases GadA/GadB enzymes and the glutamate/γ-aminobutyric acid (GABA) antiporter GadC[2]. | ||
| − | Characterization: | + | |
| + | <h2><strong>Characterization:</strong></h2> | ||
We have expressed this gene and tested its influence on the acid tolerance of E.coli MG1655-T7 RNAP (MGR). T7 RNA polymerase was integrated into the genome of E. coli MG1655(MG) to test our VerProS system. | We have expressed this gene and tested its influence on the acid tolerance of E.coli MG1655-T7 RNAP (MGR). T7 RNA polymerase was integrated into the genome of E. coli MG1655(MG) to test our VerProS system. | ||
The functional gene gadB was constructed on plasmid pET30a(+) and the plasmid was transformed into MGR. IPTG(0.2 mM) was added to induce the expression of the protein. Inoculated the MGR with pET30a(+)-gadB in 10ML LB medium,37 ℃,250rpm for 12 hours,and then 1:100 transferred it to 12.5ml medium with IPTG (0.2 mM) for 18 hours. The following is the picture of SDS-PAGE (Fig.1) which shows that target protein has been expressed successfully. | The functional gene gadB was constructed on plasmid pET30a(+) and the plasmid was transformed into MGR. IPTG(0.2 mM) was added to induce the expression of the protein. Inoculated the MGR with pET30a(+)-gadB in 10ML LB medium,37 ℃,250rpm for 12 hours,and then 1:100 transferred it to 12.5ml medium with IPTG (0.2 mM) for 18 hours. The following is the picture of SDS-PAGE (Fig.1) which shows that target protein has been expressed successfully. | ||
| − | [[File:T--SCUT China--gadB ybaS protein.jpeg| | + | <p style="text-align:center;">[[File:T--SCUT China--gadB ybaS protein.jpeg|800px]]</p> |
| − | Fig.1 The SDS-PAGE of gadB and ybaS | + | <p style="text-align:center;">Fig.1 The SDS-PAGE of gadB and ybaS</p> |
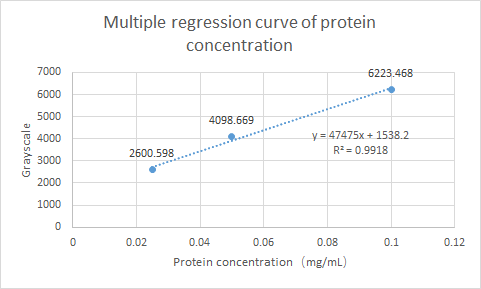
What’s more, we have tested the Protein expression of gadB. Using ImageJ for gray scale comparison, the multiple regression curve was drawn with the BSA of 0.0125m to 0.1m as the reference, as follow figure: | What’s more, we have tested the Protein expression of gadB. Using ImageJ for gray scale comparison, the multiple regression curve was drawn with the BSA of 0.0125m to 0.1m as the reference, as follow figure: | ||
| − | [[File:T--SCUT China--multiple regression curve.jpeg|500px]] | + | <p style="text-align:center">[[File:T--SCUT China--multiple regression curve.jpeg|500px]] </p> |
| − | Fig.2 Multiple regression curve of protein concentration | + | <p style="text-align:center;">Fig.2 Multiple regression curve of protein concentration </p> |
Finally, the expression of gadB was calculated as 0.0342 mg/ml. | Finally, the expression of gadB was calculated as 0.0342 mg/ml. | ||
| Line 24: | Line 24: | ||
The last, we have tested its influence on the acid tolerance of MGR. MG, MGR and MGR expressing gadB were grown overnight (about 16 h) in LBG medium of pH 7.0 at 37 °C. The cultures were then diluted to initial OD600 0.05 in 300 μL LBG medium of pH 7.0, LBG medium acidified by HCl or succinic acid to pH 4.5. Then the cultures were incubated at 37 °C in 100-well Honeycomb microplates using an automated turbidimeter (Bioscreen C, Oy Growth Curves Ab Ltd., Helsinki, Finland) for online monitoring of OD600 for 24 h. | The last, we have tested its influence on the acid tolerance of MGR. MG, MGR and MGR expressing gadB were grown overnight (about 16 h) in LBG medium of pH 7.0 at 37 °C. The cultures were then diluted to initial OD600 0.05 in 300 μL LBG medium of pH 7.0, LBG medium acidified by HCl or succinic acid to pH 4.5. Then the cultures were incubated at 37 °C in 100-well Honeycomb microplates using an automated turbidimeter (Bioscreen C, Oy Growth Curves Ab Ltd., Helsinki, Finland) for online monitoring of OD600 for 24 h. | ||
A growth assay under moderate acid stress were performed to investigate the effect of overexpression gadB or not on acid tolerance. Under moderate acid stress, the final OD600 value of strain MGR-gadB(the strain overexpressing gadB) was 44.2% higher than that of the wild type strain (MG) (Fig. 3). | A growth assay under moderate acid stress were performed to investigate the effect of overexpression gadB or not on acid tolerance. Under moderate acid stress, the final OD600 value of strain MGR-gadB(the strain overexpressing gadB) was 44.2% higher than that of the wild type strain (MG) (Fig. 3). | ||
| − | + | <p style="text-align:center;"> [[File:T - SCUT China- MR in gadB.jpeg|500px]] </p> | |
| − | Fig.3 Growth of strains MG,MGR and MGR-gadB under acid stress. | + | <p style="text-align:center;">Fig.3 Growth of strains MG,MGR and MGR-gadB under acid stress.</p> |
| − | References: | + | <h2><strong>References:</strong></h2> |
| − | [1] Feehily C, Karatzas KAG. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114:11–24. doi: 10.1111/j.1365-2672.2012.05434.x. | + | |
| + | [1] Feehily C, Karatzas KAG. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114:11–24. doi: 10.1111/j.1365-2672.2012.05434.x.<br> | ||
[2] Kanjee U, Houry WA. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol. 2013;67:65–81. 10.1146/annurev-micro-092412-155708 | [2] Kanjee U, Houry WA. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol. 2013;67:65–81. 10.1146/annurev-micro-092412-155708 | ||
| + | <h2><strong>Improvement of gadB by BUCT 2021:</strong></h2> | ||
| + | |||
| + | <html> | ||
| + | |||
| + | This year, 2021_BUCT mutated the gene gadB of 2019_SCUT ( | ||
| + | |||
| + | <a href = "https://parts.igem.org/Part:BBa_K3100017" > BBa_K3100017</a> | ||
| + | |||
| + | ) and tried to characterize GadB and GadB(mut) in different pH value. Then results of the two parts were compared. | ||
| + | Firstly, we design the experiment below to prove that we have improved part ( | ||
| + | |||
| + | <a href = "https://parts.igem.org/Part:BBa_K3100017" > BBa_K3100017</a> | ||
| + | |||
| + | ). | ||
| + | We first communicate with the leader of 2019_SCUT team to get their plasmid profile which they used to produce GadB in 2019.<br> | ||
| + | <br> | ||
| + | Below is plasmid profile of pET-30a(+)-GA-gadB,which is used by 2019_SCUT to produce GadB in 2019<br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/parts/3/3f/T--BUCT--Plasmid_map_of_pET-30a%28%2B%29-gadB%2C_which_was_used_by_2019_SCUT_team_to_produce_GadB.png"width="640px";height="30px"/><br> | ||
| + | |||
| + | Based on this plasmid, we construct a new plasmid (BBa_K3875014), which just introduce the gene gadB(mut) into pET-30a(+)-gadB to replace the gene gadB.<br> | ||
| + | |||
| + | |||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/parts/6/64/T--BUCT--Figure%2C_plasmid_profile_of_pET30-gadB_%28mut%29.png"width="640px";height="30px"/><br> | ||
| + | |||
| + | For the pET30-gadB, we decide to remove the gene gadB from E.coli B.W25113,and then going PCR amplification. After PCR amplification, pET30-gadB and a had the same cohesive end by restriction endonuclease, and then they were connected by T7 ligase. Finally, the linked plasmid was transformed into E. coli DH5a. Then we plan to run a gel to make sure that the gene is a right one. After that, in order to detect the expression of recombinant gene, the plasmid containing pET30-gadB was transformed into E. coli DH5a for fermentation (In fact, we didn't do all of these because of time constraints). | ||
| + | |||
| + | After fermentation, we extract and purify the GadB and examine its activity in different value. And based on a variety of dissertations and the experiments done by 2019_SCUT team, the result may be that GadB only maintain active in pH 4.5[2,3,4]. | ||
| + | |||
| + | For the pET30-gadB(mut), because we had successfully constructed the composite part (Lac-gadB(mut)-lac-gdhA-T1/BBa_K3875007), so we can directly remove the gene gadB(mut) from Lac-gadB(mut)-lac-gdhA-T1 (BBa_K3875007) with Primer 1 (CTTGAGACCTCCTTCTTAAAGTTAAAC) and Primer 3 (CAAGGGGTTATGCTAGTTATTGCTCTTAGTGATCGCTGAGATATTTCAGG). | ||
| + | After PCR amplification, pET30-gadB(mut) and a had the same cohesive end by restriction endonuclease, and then they were connected by T7 ligase. Finally, the linked plasmid was transformed into E. coli DH5a. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | After the Polymerase Chain Reaction, the gene run a gel. <br> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/parts/c/cd/T--BUCT%E2%80%94gel_11.png"width="640px";height="30px"/><br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | As we can see from this picture, number 9,13,15 is the proper gadB(mut) that we need.<br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/7/75/T--BUCT%E2%80%94gel_22.png"width="640px";height="30px"/><br> | ||
| + | |||
| + | In order to detect the expression of recombinant gene, the plasmid containing pET30-gadB(mut) was transformed into E. coli DH5a for fermentation (In fact, we didn't do it because of time constraints). | ||
| + | After that, we plan to purify the protein GadB (mut) and detect the protein activity of GadB in different pH value. | ||
| + | Our assumption is that our GadB (mut) would remain active at different pH, instead of just being active in pH 4.5. | ||
| + | Below is a map of our design for this experiment: | ||
| + | <img src="https://static.igem.org/mediawiki/parts/e/e9/T--BUCT--3.png"width="640px";height="30px"/><br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | [1] Sheng, L., Shen, D., Yang, W., Zhang, M., Zeng, Y., Xu, J., Deng, X., & Cheng, Y. (2017). GABA Pathway Rate-Limit Citrate Degradation in Postharvest Citrus Fruit Evidence from HB Pumelo (Citrus grandis) × Fairchild (Citrus reticulata) Hybrid Population. Journal of agricultural and food chemistry, 65(8), 1669–1676. https://doi.org/10.1021/acs.jafc.6b05237 | ||
| + | [2] Gut, H., Pennacchietti, E., John, R. A., Bossa, F., Capitani, G., De Biase, D., & Grütter, M. G. (2006). Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. The EMBO journal, 25(11), 2643–2651. https://doi.org/10.1038/sj.emboj.7601107 | ||
| + | [3] Chae, T. U., Ko, Y. S., Hwang, K. S., & Lee, S. Y. (2017). Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams. Metabolic engineering, 41, 82–91. https://doi.org/10.1016/j.ymben.2017.04.001 | ||
| + | |||
| + | </html> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 15:13, 18 October 2021
gadB (antiacid gene)
Usage and Biology:
GadB is an acid tolerant factor which play an important role in the acid tolerance of E.coli MG1655.The decarboxylation of glutamate consumes a proton, and therefore, micro‐organisms take advantage of this property to remove protons from the intracellular milieu under acidic conditions[1]. GadB is involved in the AR system, which is the glutamic acid-dependent acid resistance (GDAR) system, consisting of the homologous inducible glutamic acid decarboxylases GadA/GadB enzymes and the glutamate/γ-aminobutyric acid (GABA) antiporter GadC[2].
Characterization:
We have expressed this gene and tested its influence on the acid tolerance of E.coli MG1655-T7 RNAP (MGR). T7 RNA polymerase was integrated into the genome of E. coli MG1655(MG) to test our VerProS system.
The functional gene gadB was constructed on plasmid pET30a(+) and the plasmid was transformed into MGR. IPTG(0.2 mM) was added to induce the expression of the protein. Inoculated the MGR with pET30a(+)-gadB in 10ML LB medium,37 ℃,250rpm for 12 hours,and then 1:100 transferred it to 12.5ml medium with IPTG (0.2 mM) for 18 hours. The following is the picture of SDS-PAGE (Fig.1) which shows that target protein has been expressed successfully.
Fig.1 The SDS-PAGE of gadB and ybaS
What’s more, we have tested the Protein expression of gadB. Using ImageJ for gray scale comparison, the multiple regression curve was drawn with the BSA of 0.0125m to 0.1m as the reference, as follow figure:
Fig.2 Multiple regression curve of protein concentration
Finally, the expression of gadB was calculated as 0.0342 mg/ml.
The last, we have tested its influence on the acid tolerance of MGR. MG, MGR and MGR expressing gadB were grown overnight (about 16 h) in LBG medium of pH 7.0 at 37 °C. The cultures were then diluted to initial OD600 0.05 in 300 μL LBG medium of pH 7.0, LBG medium acidified by HCl or succinic acid to pH 4.5. Then the cultures were incubated at 37 °C in 100-well Honeycomb microplates using an automated turbidimeter (Bioscreen C, Oy Growth Curves Ab Ltd., Helsinki, Finland) for online monitoring of OD600 for 24 h. A growth assay under moderate acid stress were performed to investigate the effect of overexpression gadB or not on acid tolerance. Under moderate acid stress, the final OD600 value of strain MGR-gadB(the strain overexpressing gadB) was 44.2% higher than that of the wild type strain (MG) (Fig. 3).
Fig.3 Growth of strains MG,MGR and MGR-gadB under acid stress.
References:
[1] Feehily C, Karatzas KAG. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J Appl Microbiol. 2013;114:11–24. doi: 10.1111/j.1365-2672.2012.05434.x.
[2] Kanjee U, Houry WA. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol. 2013;67:65–81. 10.1146/annurev-micro-092412-155708
Improvement of gadB by BUCT 2021:
This year, 2021_BUCT mutated the gene gadB of 2019_SCUT (
BBa_K3100017
) and tried to characterize GadB and GadB(mut) in different pH value. Then results of the two parts were compared.
Firstly, we design the experiment below to prove that we have improved part (
BBa_K3100017
).
We first communicate with the leader of 2019_SCUT team to get their plasmid profile which they used to produce GadB in 2019.
Below is plasmid profile of pET-30a(+)-GA-gadB,which is used by 2019_SCUT to produce GadB in 2019

Based on this plasmid, we construct a new plasmid (BBa_K3875014), which just introduce the gene gadB(mut) into pET-30a(+)-gadB to replace the gene gadB.

For the pET30-gadB, we decide to remove the gene gadB from E.coli B.W25113,and then going PCR amplification. After PCR amplification, pET30-gadB and a had the same cohesive end by restriction endonuclease, and then they were connected by T7 ligase. Finally, the linked plasmid was transformed into E. coli DH5a. Then we plan to run a gel to make sure that the gene is a right one. After that, in order to detect the expression of recombinant gene, the plasmid containing pET30-gadB was transformed into E. coli DH5a for fermentation (In fact, we didn't do all of these because of time constraints).
After fermentation, we extract and purify the GadB and examine its activity in different value. And based on a variety of dissertations and the experiments done by 2019_SCUT team, the result may be that GadB only maintain active in pH 4.5[2,3,4].
For the pET30-gadB(mut), because we had successfully constructed the composite part (Lac-gadB(mut)-lac-gdhA-T1/BBa_K3875007), so we can directly remove the gene gadB(mut) from Lac-gadB(mut)-lac-gdhA-T1 (BBa_K3875007) with Primer 1 (CTTGAGACCTCCTTCTTAAAGTTAAAC) and Primer 3 (CAAGGGGTTATGCTAGTTATTGCTCTTAGTGATCGCTGAGATATTTCAGG).
After PCR amplification, pET30-gadB(mut) and a had the same cohesive end by restriction endonuclease, and then they were connected by T7 ligase. Finally, the linked plasmid was transformed into E. coli DH5a.
After the Polymerase Chain Reaction, the gene run a gel.

As we can see from this picture, number 9,13,15 is the proper gadB(mut) that we need.

In order to detect the expression of recombinant gene, the plasmid containing pET30-gadB(mut) was transformed into E. coli DH5a for fermentation (In fact, we didn't do it because of time constraints).
After that, we plan to purify the protein GadB (mut) and detect the protein activity of GadB in different pH value.
Our assumption is that our GadB (mut) would remain active at different pH, instead of just being active in pH 4.5.
Below is a map of our design for this experiment:

[1] Sheng, L., Shen, D., Yang, W., Zhang, M., Zeng, Y., Xu, J., Deng, X., & Cheng, Y. (2017). GABA Pathway Rate-Limit Citrate Degradation in Postharvest Citrus Fruit Evidence from HB Pumelo (Citrus grandis) × Fairchild (Citrus reticulata) Hybrid Population. Journal of agricultural and food chemistry, 65(8), 1669–1676. https://doi.org/10.1021/acs.jafc.6b05237
[2] Gut, H., Pennacchietti, E., John, R. A., Bossa, F., Capitani, G., De Biase, D., & Grütter, M. G. (2006). Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB. The EMBO journal, 25(11), 2643–2651. https://doi.org/10.1038/sj.emboj.7601107
[3] Chae, T. U., Ko, Y. S., Hwang, K. S., & Lee, S. Y. (2017). Metabolic engineering of Escherichia coli for the production of four-, five- and six-carbon lactams. Metabolic engineering, 41, 82–91. https://doi.org/10.1016/j.ymben.2017.04.001
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 988
Illegal PstI site found at 440
Illegal PstI site found at 1360 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 988
Illegal PstI site found at 440
Illegal PstI site found at 1360 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 988
- 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 988
Illegal PstI site found at 440
Illegal PstI site found at 1360 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 988
Illegal PstI site found at 440
Illegal PstI site found at 1360
Illegal AgeI site found at 700 - 1000COMPATIBLE WITH RFC[1000]