Difference between revisions of "Part:BBa K3771094"
Karin100699 (Talk | contribs) |
|||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3771094 short</partinfo> | <partinfo>BBa_K3771094 short</partinfo> | ||
| − | + | <br><b style="font-size:1.3rem">Description</b> | |
| + | <br> | ||
| + | |||
| + | <br>This composite part consists of promoter <i>P<sub>soxS</sub></i> (BBa_K3771048) and the cs gene (BBa_K3771012), which encodes L-cysteine sulfonic acid synthase (CS). It is used to express CS to produce taurine under oxidative stress. | ||
| + | The <i>soxS</i> gene is one component of the <i>soxRS</i> regulon, which is important for <i>E. coli</i> to sense and respond to the oxidants. The <i>soxS</i> promoter (<i>P<sub>soxS</sub></i>) is stimulated by oxidative stress, leading to the expression of the downstream gene. The L-cysteine sulfonic acid synthase (CS) is an enzyme that catalyzes the conversion of O-phospho-L-serine and sulfite into L-cysteine sulfonic acid (L-cysteate) and phosphate. In order to use anti-polyhistidine-tag antibodies to detect the production of CS by western blot, we add 6xHis-tag at the C-terminal of CS. | ||
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | <!-- --> | ||
| + | <br><b style="font-size:1.3rem">Usage and Biology</b> | ||
| + | <br> | ||
| + | |||
| + | <br>This composite part was ligated with the pSAA vector and transformed into <i>E. coli</i>. We conducted colony PCR to verify whether <i>E. coli</i> uptake the correct plasmid. The size of the PCR product was as expected.The part has been confirmed by sequencing and has no mutations. | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <html><div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://2021.igem.org/wiki/images/4/41/T--NCKU_Tainan--soxR-sfGFP_Colony_PCR.png" style="width:35%;"> | ||
| + | </div></html> | ||
| + | <p align="center"> Fig.1. The electrophoresis result of colony PCR. M: Marker; Lane 1: pSAA-<i>P<sub>soxS</sub></i>-<i>sfgfp</i> (2691 bp); Lane 2, 3: pSAA-<i>P<sub>soxS</sub>-cs-6xHis</i> (3240 bp). | ||
| + | </p> | ||
| + | |||
| + | <br>The promoter strength under oxidative stress of <i>P<sub>soxS</sub></i> with its activator, SoxR, is determined by the expression level of sfGFP, which is their reporter. While using hydrogen peroxide (H<sub>2</sub>O<sub>2</sub>) as the inducer, the intensity of soxR-<i>P<sub>soxS</sub></i> shows no significant difference under different concentrations of hydrogen peroxide (Fig.2). However, soxR-<i>P<sub>soxS</sub></i> is well induced by using paraquat (PQ), which is a commonly used agent to induce oxidative stress for bacteria (Fig.3 & 4). | ||
| + | <br> | ||
| + | |||
| + | <html><div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/1/17/T--NCKU_Tainan--sfGFP_Expression_%28soxR-PsoxS%29.png" style="width:35%;"> | ||
| + | </div></html> | ||
| + | <p align="center"> Fig.2. Relative fluorescence intensity of soxR-<i>P<sub>soxS</sub></i> after 4.5-hour incubation with hydrogen peroxide in various concentrations. | ||
| + | </p> | ||
| + | |||
| + | <html><div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/3/33/T--NCKU_Tainan--Del_Low_sfGFP_Expression_%28soxR-PsoxS%29.png" style="width:35%;"> | ||
| + | </div></html> | ||
| + | <p align="center"> Fig.3. Relative fluorescence intensity of soxR-<i>P<sub>soxS</sub></i> after 4.5-hour incubation with hydrogen peroxide in low concentrations. | ||
| + | </p> | ||
| + | |||
| + | <html><div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/0/0c/T--NCKU_Tainan--Del_High_sfGFP_Expression_%28soxR-PsoxS%29.png" style="width:35%;"> | ||
| + | </div></html> | ||
| + | <p align="center"> Fig.4. Relative fluorescence intensity of soxR-<i>P<sub>soxS</sub></i> after 4.5-hour incubation with hydrogen peroxide in high concentrations. | ||
| + | </p> | ||
| + | |||
| + | <br>We compared the relative fluorescence intensity of sfGFP expressed by different promoters under the stimulation by different concentrations of paraquat. As expected, sfGFP expression level of J23100 shows no difference between the groups with and without paraquat. <i>P<sub>soxS</sub></i> with its activator, SoxR (BBa_K37710XX), is highly activated under oxidative stress and has the highest gene expression level compared to J23100, <i>P<sub>katG</sub></i> (BBa_K37710XX), and <i>P<sub>soxS</sub></i> (BBa_K37710XX) solely (Fig.5). Moreover, we compared the efficiency of our design with (BBa_K37710XX), in which the concept came from the 2018 Imperial College London iGEM team. Different from our design, they used a mutant <i>P<sub>soxS</sub></i> promoter with RiboJ (BBa_K2862010) and expressed SoxR by J23101 promoter. However, the sfGFP expression by BBa_K2862010 was quite low (Fig.6). | ||
| + | <br> | ||
| + | |||
| + | <html><div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/c/c2/T--NCKU_Tainan--sfGFP_Expression_%28PQ_%404.5h%29.png" style="width:35%;"> | ||
| + | </div></html> | ||
| + | <p align="center"> Fig.5. Comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in high concentrations. | ||
| + | </p> | ||
| + | |||
| + | <html><div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/0/0d/T--NCKU_Tainan--Low_sfGFP_Expression_%28PQ_%404.5h%29.png" style="width:35%;"> | ||
| + | </div></html> | ||
| + | <p align="center"> Fig.6. Comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in low concentrations. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <br><b style="font-size:1.3rem">References</b> | ||
| + | <br> | ||
| + | |||
| + | <br>1. Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19(3):109-114. doi:10.1016/s0167-7799(00)01542-0 | ||
| + | <br> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 02:48, 18 October 2021
PsoxS-CS-6xHis
Description
This composite part consists of promoter PsoxS (BBa_K3771048) and the cs gene (BBa_K3771012), which encodes L-cysteine sulfonic acid synthase (CS). It is used to express CS to produce taurine under oxidative stress.
The soxS gene is one component of the soxRS regulon, which is important for E. coli to sense and respond to the oxidants. The soxS promoter (PsoxS) is stimulated by oxidative stress, leading to the expression of the downstream gene. The L-cysteine sulfonic acid synthase (CS) is an enzyme that catalyzes the conversion of O-phospho-L-serine and sulfite into L-cysteine sulfonic acid (L-cysteate) and phosphate. In order to use anti-polyhistidine-tag antibodies to detect the production of CS by western blot, we add 6xHis-tag at the C-terminal of CS.
Usage and Biology
This composite part was ligated with the pSAA vector and transformed into E. coli. We conducted colony PCR to verify whether E. coli uptake the correct plasmid. The size of the PCR product was as expected.The part has been confirmed by sequencing and has no mutations.

Fig.1. The electrophoresis result of colony PCR. M: Marker; Lane 1: pSAA-PsoxS-sfgfp (2691 bp); Lane 2, 3: pSAA-PsoxS-cs-6xHis (3240 bp).
The promoter strength under oxidative stress of PsoxS with its activator, SoxR, is determined by the expression level of sfGFP, which is their reporter. While using hydrogen peroxide (H2O2) as the inducer, the intensity of soxR-PsoxS shows no significant difference under different concentrations of hydrogen peroxide (Fig.2). However, soxR-PsoxS is well induced by using paraquat (PQ), which is a commonly used agent to induce oxidative stress for bacteria (Fig.3 & 4).

Fig.2. Relative fluorescence intensity of soxR-PsoxS after 4.5-hour incubation with hydrogen peroxide in various concentrations.

Fig.3. Relative fluorescence intensity of soxR-PsoxS after 4.5-hour incubation with hydrogen peroxide in low concentrations.
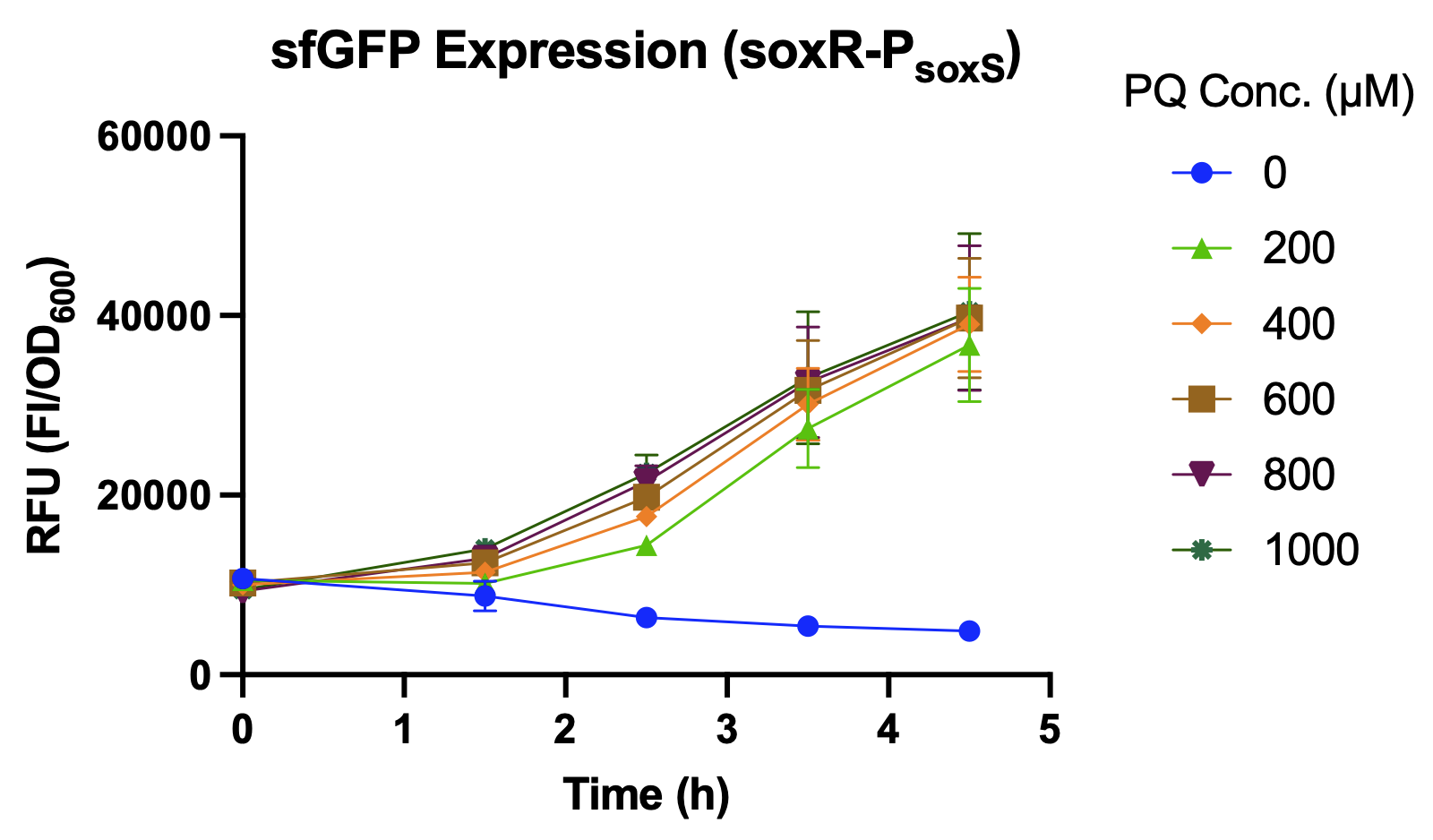
Fig.4. Relative fluorescence intensity of soxR-PsoxS after 4.5-hour incubation with hydrogen peroxide in high concentrations.
We compared the relative fluorescence intensity of sfGFP expressed by different promoters under the stimulation by different concentrations of paraquat. As expected, sfGFP expression level of J23100 shows no difference between the groups with and without paraquat. PsoxS with its activator, SoxR (BBa_K37710XX), is highly activated under oxidative stress and has the highest gene expression level compared to J23100, PkatG (BBa_K37710XX), and PsoxS (BBa_K37710XX) solely (Fig.5). Moreover, we compared the efficiency of our design with (BBa_K37710XX), in which the concept came from the 2018 Imperial College London iGEM team. Different from our design, they used a mutant PsoxS promoter with RiboJ (BBa_K2862010) and expressed SoxR by J23101 promoter. However, the sfGFP expression by BBa_K2862010 was quite low (Fig.6).

Fig.5. Comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in high concentrations.

Fig.6. Comparison of different promoter strengths by relative fluorescence intensity after 4.5-hour incubation with paraquat in low concentrations.
References
1. Pomposiello PJ, Demple B. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 2001;19(3):109-114. doi:10.1016/s0167-7799(00)01542-0
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 604
