Difference between revisions of "Part:BBa K808027"
| (3 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K808027 short</partinfo> | <partinfo>BBa_K808027 short</partinfo> | ||
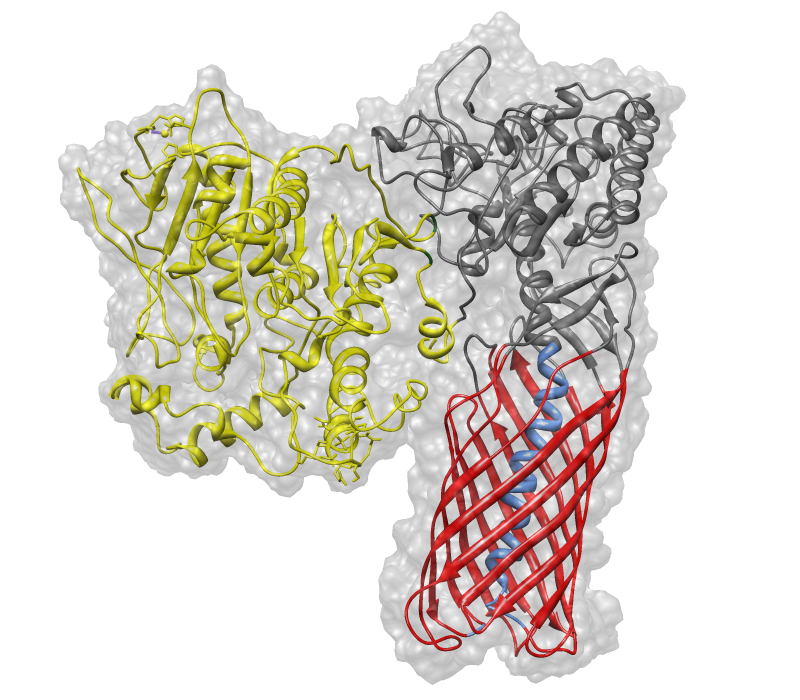
| − | + | EstA is a membrane bound but inactive esterase from ''Pseudomonas aeruginosa''. It can be used (unlike https://parts.igem.org/Part:BBa_K633001 BBa_K633001) for the cell surface expression of enzymes. Similiar to OmpA it allows to perform bacterial display of e.g. hydrolases like it has already been shown by Becker, Theile, Heppeler & Michalczyk, 2005. We used EstA to create our chimeric protein [https://parts.igem.org/Part:BBa_K808030 BBa_K808030] (Figure 1). | |
| + | |||
| + | |||
| + | [[Image:pnb_EstA_ohne_mem.png|400px|center|thumb|Figure 1:Visualization of [https://parts.igem.org/Part:BBa_K808030 BBa_K808030]: EstA (red & grey) anchoring a second protein (pNB-Est13 in yellow) into a biological membrane]] | ||
| + | |||
| + | |||
| + | =IISER_BHOPAL 2021= | ||
| + | ===Cold adapted Esterase=== | ||
| + | A cold-adapted esterase was extracted from psychrotrophic bacterium Pseudoalteromonas sp. strain 643A, which is present in the alimentary tract of Antarctic krill Euphasia superba Dana. This protein consists of 206 amino acids residue with a molecular mass of 23,036 Da and belongs to the member of the GDSL-lipolytic enzymes family.1 | ||
| + | |||
| + | ===References=== | ||
| + | Cieśliński, H., Białkowska, A. M., Długołęcka, A., Daroch, M., Tkaczuk, K. L., Kalinowska, H., Kur, J., & Turkiewicz, M. (2007). A cold-adapted esterase from psychrotrophic Pseudoalteromas sp. strain 643A. Archives of Microbiology, 188(1), 27–36. https://doi.org/10.1007/s00203-007-0220-2 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| − | |||
<!-- --> | <!-- --> | ||
Latest revision as of 14:34, 9 October 2021
EstA : inactive fusion protein for bacterial surface expression
EstA is a membrane bound but inactive esterase from Pseudomonas aeruginosa. It can be used (unlike https://parts.igem.org/Part:BBa_K633001 BBa_K633001) for the cell surface expression of enzymes. Similiar to OmpA it allows to perform bacterial display of e.g. hydrolases like it has already been shown by Becker, Theile, Heppeler & Michalczyk, 2005. We used EstA to create our chimeric protein BBa_K808030 (Figure 1).
IISER_BHOPAL 2021
Cold adapted Esterase
A cold-adapted esterase was extracted from psychrotrophic bacterium Pseudoalteromonas sp. strain 643A, which is present in the alimentary tract of Antarctic krill Euphasia superba Dana. This protein consists of 206 amino acids residue with a molecular mass of 23,036 Da and belongs to the member of the GDSL-lipolytic enzymes family.1
References
Cieśliński, H., Białkowska, A. M., Długołęcka, A., Daroch, M., Tkaczuk, K. L., Kalinowska, H., Kur, J., & Turkiewicz, M. (2007). A cold-adapted esterase from psychrotrophic Pseudoalteromas sp. strain 643A. Archives of Microbiology, 188(1), 27–36. https://doi.org/10.1007/s00203-007-0220-2
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 97
Illegal NgoMIV site found at 235
Illegal NgoMIV site found at 775
Illegal NgoMIV site found at 1144 - 1000COMPATIBLE WITH RFC[1000]
