Difference between revisions of "Part:BBa K3610046"
(→Confocal Fluorescence Microscopy) |
(→Characterization) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 161: | Line 161: | ||
[[File:T--UZurich--FACs.png|600px|thumb|none|Figure 6: Left: single biological replicates. right: pooled samples. With the exception of the construct with eCORE, cells transfected with our constructs showed considerably higher overall fluorescence intensities than the negative control.]] | [[File:T--UZurich--FACs.png|600px|thumb|none|Figure 6: Left: single biological replicates. right: pooled samples. With the exception of the construct with eCORE, cells transfected with our constructs showed considerably higher overall fluorescence intensities than the negative control.]] | ||
| + | |||
===Conclusion=== | ===Conclusion=== | ||
| − | Multiple experiments suggest that expression levels of this construct in <i>S. cerevisiae</i> are very weak and | + | Multiple experiments suggest that expression levels of this construct in <i>S. cerevisiae</i> are very weak and we did not observe any fluorescence overlapping with the membrane stain. |
| + | |||
<!-- --> | <!-- --> | ||
Latest revision as of 23:58, 27 October 2020
CORE ectodomain / YFP
This part contains the ectodomain of the plant cell surface receptor CORE from S. lycopersicum fused to a yellow fluorescent protein. This part lacks the natural N-terminal signal sequence but instead uses the signal sequence from the alpha-Factor from yeast.
Usage and Biology
CORE
The cold shock protein receptor (CORE) is a plant pattern recognition receptor (PRR) and as such activates host innate immunity through detection of pathogen-associated molecular patterns (PAMPs). CORE is a leucine-rich repeat receptor-like kinase with 22 LRRs, there is a 6 amino acid insert at LRR 11. It consists of an extracellular domain that perceives an epitope, csp22, from the highly conserved nucleic acid binding motif RNP-1 of bacterial cold-shock proteins (CSPs), which are highly abundant proteins found in the cytosol of bacteria. Further domains are a single pass transmembrane domain and an intracellular kinase domain (The sequence encoding the kinase domain is not in this part). Interaction of CORE with brassinosteroid-associated kinase (BAK)1 is necessary for inducing an immune response in the plant. The dimerization of CORE and BAK1 depends on the csp22, the ligand of CORE. The function of CORE in S. lycopersicum has been confirmed by expressing the receptor in A. thaliana, which made the plant responsive to csp22, a PAMP that is otherwise not perceived by PRRs from A. thaliana.
CORE with YFP
In this sequence, the C-terminal domain entailing the intracellular kinase domain was replaced with the sequence coding for the yellow fluorescent protein venus, while the ectodomain and the transmembrane domain, including the juxtamembrane domain were kept. Additionally, a signal sequence native to S. cerevisiae was fused to the N-terminal sequence, which does not contain the native signal peptide. This way, the protein can be integrated into the membrane during translation and the expression can be observed as with the receptor protein, the YFP (Exλ : 515 nm, Emλ : 528 nm) gets translated as well.
Characterization
Expression of CORE ectodomain / YFP in S. cerevisiae
In a first step we inserted the single fragments making up this part into a plasmid with a gentamycin-3-acetyltransferase gene and transformed E. coli (DH10alpha) with the plasmids for amplification. In the next step we assembled the fragments in a plasmid with a spectinomycin acetyltransferase and amplified the plasmids again in the same E. coli strain. For this step we applied the techniques of Golden Gate Cloning to get the fragments in the right order into the plasmid. The restriction enzyme we chose was BsaI. For expressing this part consisting of YFP and the receptor protein, we initially intended to use promoters of different strengths to get more quantitative data. Finally, we got the construct in a plasmid with a truncated version of the ADH1 promoter from S. cerevisiae. For termination, this part has the terminator sequence of the enolase 2 protein from S. cerevisiae. The plasmid also contained the TRP1 gene, which encodes phosphoribosylanthranilate isomerase, an enzyme that catalyzes the third step in tryptophan biosynthesis. This enabled us to use the same plasmid for expression in S. cerevisiae. We prepared a medium containing YNB and free amino acids, without tryptophan. S. cerevisiae cells (AP4) were transfected with the plasmid and then plated on the selective medium.
Confocal Fluorescence Microscopy
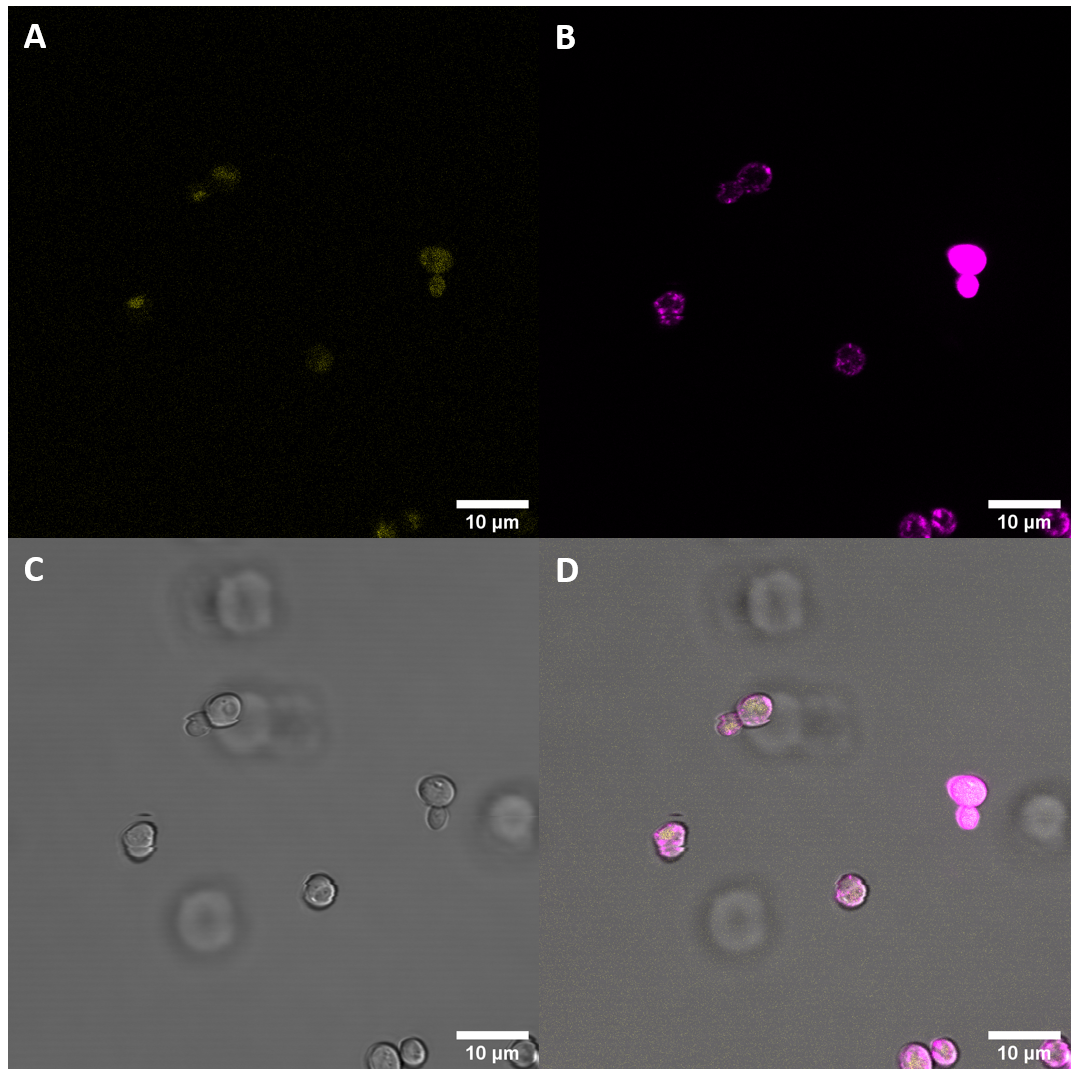
After successful transformation of yeast cells we checked for expression of the protein under a confocal microscope. If expression of YFP (λEx = 515 nm, λEx = 528 nm) can clearly be observed, it is reasonable to assume that the receptor domain is expressed as well, as the YFP is fused to the receptor protein. Expression of the construct was confirmed. We failed, however, to confirm localization at the cell membrane.
Initial imaging did not reveal increased fluorescence at the excitation and emission wavelengths for YFP. As localization at the cell membrane was something we were particularily interested in, we repeated the confocal microscopy step with an additional membrane stain. The cell membrane was stained with fm4-64, which fluoresces strongly after binding to the cell membrane ((λEX = 515nm and λEM = 640nm). The binding of the dye is happening rapidly and it is also reversible. If the time spent between staining and imaging is too long, then the dye will be taken up by the organism and stored in the vacuole. Imaging with a confocal microscope for YFP and the fm4-64 stain shows the spatial overlap of the red fluorescence of the stain and the yellow fluorescence of the protein fused to the receptors.
Fluorescenc Microscopy showed, that CORE does not get expressed very well in S. cerevisiae. There is also no observeable localization at the plasma membrane.
Spectrometry
In addition to analyzing the cells with a microscope, we conducted a fluorescence assay with a plate reader. We conducted this experiment for multiple receptors at the same time. This way we were able to compare the expression levels of differnt versions of the BAK1 receptor. For each receptor we tried to isolate three different biological samples, however, not all cells grew. Ultimately, we only had two samples for the following S. cerevisiae cells: untransformed (Control), transformed with BAK1 ectodomain fused to YFP (eBAK) and the CORE ectodomain fused to YFP (eCORE). For the BAK1 with and without the native signal peptide fused to YFP (BAK+ and BAK-) and the EFR ectodomain fused to YFP (eEFR), we had samples from three different colonies. For each biological replicate, the optical density at absorbance of 600 nm (OD600) and the fluorescence levels were measured three times.
| measured OD600 values (OD) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | |||||||
| Blank | 0,08200000226 | 0,08200000226 | 0,08389999717 | ||||||
| Control | 0,3806000054 | 0,3747999966 | 0,4221999943 | 0,1316999942 | 0,131400004 | 0,1176000014 | |||
| BAK+ | 0,4943000078 | 0,4638999999 | 0,4514000118 | 0,5781000257 | 0,5253999829 | 0,5799999833 | 0,2615999877 | 0,2171999961 | 0,2011999935 |
| BAK- | 1,417099953 | 1,365499973 | 1,368899941 | 0,6305999756 | 0,5633999705 | 0,6216999888 | 0,896600008 | 0,7882999778 | 0,8032000065 |
| eBAK | 1,009699941 | 0,8404999971 | 0,8934999704 | 0,2653000057 | 0,2368000001 | 0,2592999935 | |||
| eCORE | 1,021499991 | 0,8616999984 | 0,9178000093 | 0,826300025 | 0,6888999939 | 0,7401999831 | |||
| eEFR | 1,379699945 | 1,322700024 | 1,333500028 | 1,035899997 | 1,014000058 | 0,9526000023 | 0,4860999882 | 0,3797000051 | 0,3829999864 |
The following settings were applied for fluorescence measurements:
| Mode: | Fluorescence Top Reading |
| Excitation Wavelength: | 485 nm |
| Emission Wavelength: | 535 nm |
| Excitation Bandwidth: | 20 nm |
| Emission Bandwidth: | 25 nm |
| Temperature: | 22.3°C |
| Fluorescence Top Reading (FTR) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 3 | |||||||
| Blank | 1297 | 1282 | 1322 | ||||||
| Control | 2684 | 2474 | 2634 | 1852 | 1792 | 1750 | |||
| BAK+ | 3038 | 2813 | 2760 | 2836 | 2493 | 2788 | 2084 | 2072 | 2067 |
| BAK- | 35794 | 30319 | 31424 | 10792 | 9097 | 10517 | 22609 | 20227 | 21220 |
| eBAK | 26455 | 19828 | 21613 | 6614 | 5507 | 6229 | |||
| eCORE | 10709 | 8382 | 9339 | 8957 | 7062 | 7735 | |||
| eEFR | 43125 | 37782 | 39589 | 25641 | 24668 | 22517 | 12410 | 9054 | 9027 |
After measurement of the optical density and the fluorescence, the data were blank corrected (the average of the three blank measurements was subtracted from each measurement value).
The average of each of the three (or two) samples was calculated. From these values, the average was taken again.
After this step, we normalized the fluorescent output for OD600 (FTR/OD). The results of these calculations are displayed in the table below.
| Control | BAK+ | BAK- | eBAK | eCORE | eEFR |
|---|---|---|---|---|---|
| 4185,221063 | 9731,614266 | 26067,19254 | 28118,24739 | 3712,946478 | 23379,84399 |
If we set the values for the Control to 1 (Control = 1), then we get the fluorescence levels relative to the control, which is again diplayed in the table below.
| Control | eCORE | eEFR | BAK- | BAK+ | eBAK |
|---|---|---|---|---|---|
| 1 | 0,8871565975 | 5,586286516 | 6,228390841 | 2,325233033 | 6,718461693 |
Flow Cytometry
It has been important to us to examine a sample with different approaches simultaneously, which is why we were eager to also measure fluorescence intensity by flow cytometry. In a first phase, 100,000 cells were measured from each biological replicate (488/530 FITC channel in a BD FACSCanto II flow cytometer). In the next phase, the biological replicates for each construct were pooled together and 200,000 cells from each sample were measured.
Conclusion
Multiple experiments suggest that expression levels of this construct in S. cerevisiae are very weak and we did not observe any fluorescence overlapping with the membrane stain.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1740
Illegal BamHI site found at 373
Illegal BamHI site found at 1765
Illegal BamHI site found at 2117 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]