Difference between revisions of "Part:BBa K3645013"
Teddy Huang (Talk | contribs) (→Peking iGEM 2020's documentation) |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 17: | Line 17: | ||
<partinfo>BBa_K3645013 parameters</partinfo> | <partinfo>BBa_K3645013 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | == Peking iGEM 2020's documentation == | ||
| + | <p>David Liu’s lab created the first base editor in 2016 (Komor et al., 2016) and since then has been trying to expand | ||
| + | their precision editing capabilities. Base editors make specific DNA base changes and consist of a catalytically | ||
| + | impaired Cas protein (dCas or Cas nickase) fused to a DNA-modifying enzyme, in this case a deaminase. Base changes | ||
| + | from C•G-to-T•A are mediated by cytosine base editors (CBEs) and base changes from A•T-to-G•C are mediated by | ||
| + | adenine base editors (ABEs). How does this work? Through molecular biology teamwork. The guide RNA (gRNA) specifies | ||
| + | the editing target site on the DNA, the Cas domain directs the modifying enzyme to the target site, and the | ||
| + | deaminase induces the DNA base change without a DNA double-strand break. But base editors aren’t perfect. They may | ||
| + | be slow, can only target certain sites, or make only a subset of base substitutions. (addgene blog by Susanna | ||
| + | Bachle) </p> | ||
| + | <p>We used the existing plasmids for enzyme digestion and ligation, and ePCR was added to the BioBrick connector. After | ||
| + | multiple rounds of splicing and assembly, we obtained the ABE and CBE we needed. The schematic diagrams are as | ||
| + | follows:</p> | ||
| + | <h1>CBE</h1> | ||
| + | <p>Until 2016, precise single base changes were only possible through exploiting the homology-directed repair (HDR) | ||
| + | pathway which occurs in cells at low frequencies and relies on the efficient delivery of donor DNA to facilitate | ||
| + | repair. Since the development of CRISPR-mediated base editing (BE), these types of repairs can now be done more | ||
| + | efficiently than before. A base editor precisely changes a single base with an efficiency typically ranging from | ||
| + | 2575%, while the success of precise change via HDR limited to 0-5%. This blog post covers a brief review of | ||
| + | different basic BE technologies and their adaptation for plant genome editing. (addgene blog by Guest Blogger) </p> | ||
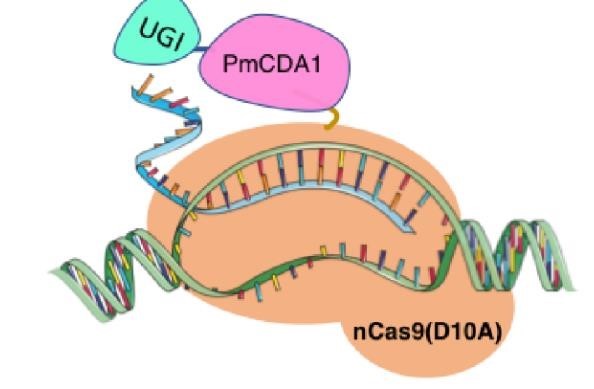
| + | <p>In 2016, two independent groups (komor et al., 2016 and Nishida et al., 2016) invented CRISPR base editor by linking | ||
| + | cytosine deaminase with cas9 cleavage enzyme (ncas9), thus achieving accurate and efficient base rewriting in the | ||
| + | genome. Ncas9 creates a gap in DNA by cutting only one single strand, thus greatly reducing the possibility of | ||
| + | harmful insertion deletion. After binding with DNA, CBE deamination of target cytosine (C) into uracil (U) base. | ||
| + | Later, the resulting U•G pairs were repaired through the cell mismatch repair mechanism to convert the original C•G | ||
| + | pair into T•A, or reduced to the original C•G through the uracil glycosylase mediated base excision repair. The | ||
| + | presence of UGI minimizes the second result and increases the production of required T•A base pairs. </p> | ||
| + | https://2020.igem.org/wiki/images/b/b4/T--Peking--CBE-5.jpg | ||
| + | <h6>(addgene blog by Guest Blogger)</h6> | ||
| + | https://2020.igem.org/wiki/images/2/2f/T--Peking--CBE-1.png | ||
| + | https://2020.igem.org/wiki/images/e/e6/T--Peking--CBE-2.png | ||
| + | https://2020.igem.org/wiki/images/6/64/T--Peking--CBE-3.png | ||
| + | https://2020.igem.org/wiki/images/6/6c/T--Peking--CBE-4.png | ||
Latest revision as of 21:32, 27 October 2020
CBE
Contains a full circuit with pBAD as a regulatory promoter. Need sgRNA expression for the base editor to function.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1205
Illegal NheI site found at 2338 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 6014
Illegal BamHI site found at 1144
Illegal BamHI site found at 4617
Illegal XhoI site found at 5623 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 979
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 961
Peking iGEM 2020's documentation
David Liu’s lab created the first base editor in 2016 (Komor et al., 2016) and since then has been trying to expand their precision editing capabilities. Base editors make specific DNA base changes and consist of a catalytically impaired Cas protein (dCas or Cas nickase) fused to a DNA-modifying enzyme, in this case a deaminase. Base changes from C•G-to-T•A are mediated by cytosine base editors (CBEs) and base changes from A•T-to-G•C are mediated by adenine base editors (ABEs). How does this work? Through molecular biology teamwork. The guide RNA (gRNA) specifies the editing target site on the DNA, the Cas domain directs the modifying enzyme to the target site, and the deaminase induces the DNA base change without a DNA double-strand break. But base editors aren’t perfect. They may be slow, can only target certain sites, or make only a subset of base substitutions. (addgene blog by Susanna Bachle)
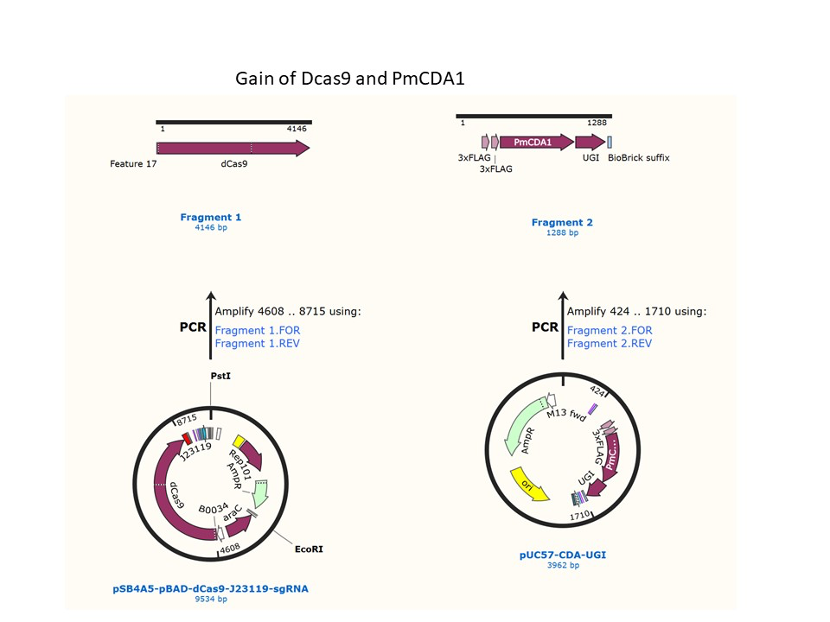
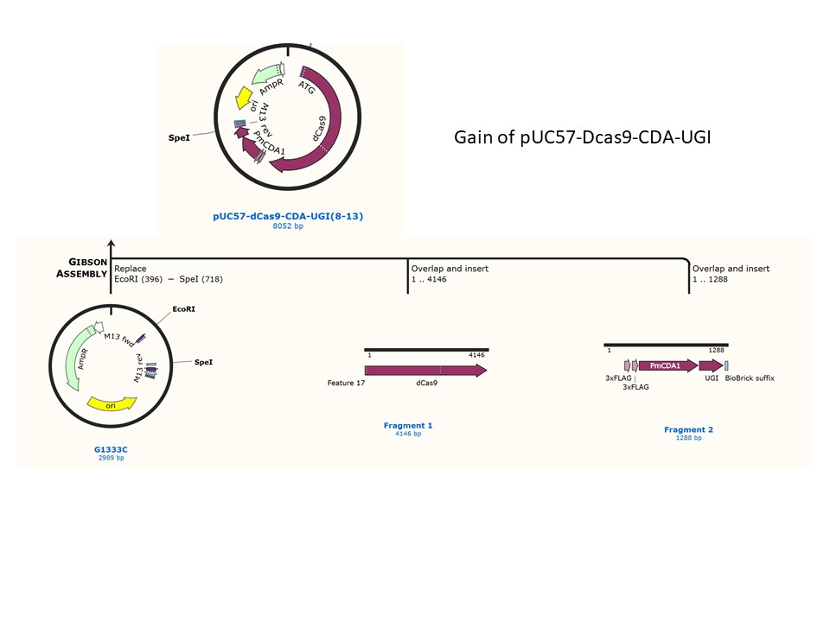
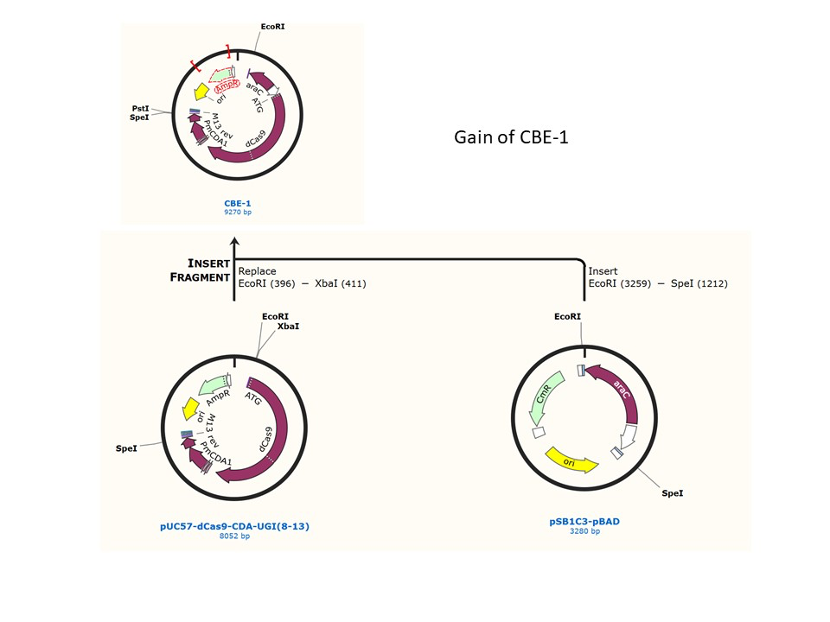
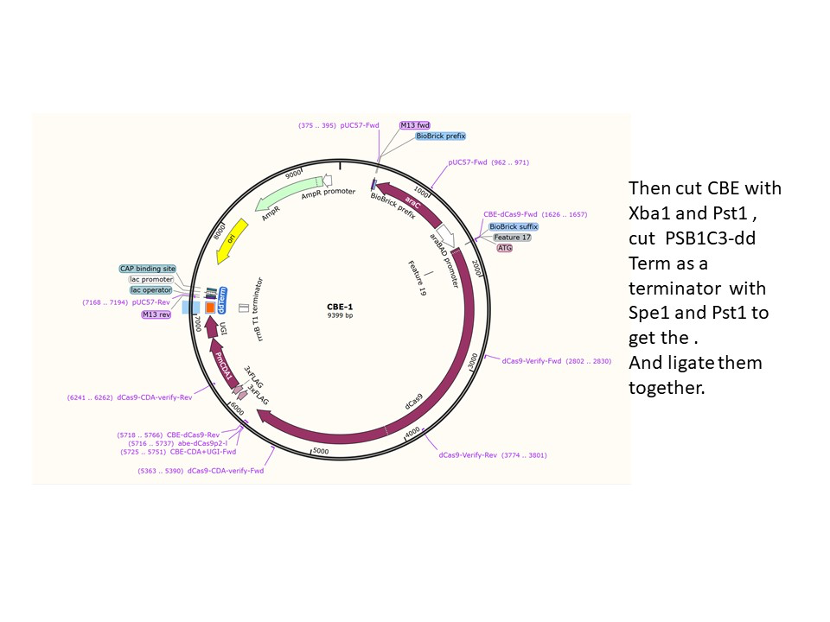
We used the existing plasmids for enzyme digestion and ligation, and ePCR was added to the BioBrick connector. After multiple rounds of splicing and assembly, we obtained the ABE and CBE we needed. The schematic diagrams are as follows:
CBE
Until 2016, precise single base changes were only possible through exploiting the homology-directed repair (HDR) pathway which occurs in cells at low frequencies and relies on the efficient delivery of donor DNA to facilitate repair. Since the development of CRISPR-mediated base editing (BE), these types of repairs can now be done more efficiently than before. A base editor precisely changes a single base with an efficiency typically ranging from 2575%, while the success of precise change via HDR limited to 0-5%. This blog post covers a brief review of different basic BE technologies and their adaptation for plant genome editing. (addgene blog by Guest Blogger)
In 2016, two independent groups (komor et al., 2016 and Nishida et al., 2016) invented CRISPR base editor by linking cytosine deaminase with cas9 cleavage enzyme (ncas9), thus achieving accurate and efficient base rewriting in the genome. Ncas9 creates a gap in DNA by cutting only one single strand, thus greatly reducing the possibility of harmful insertion deletion. After binding with DNA, CBE deamination of target cytosine (C) into uracil (U) base. Later, the resulting U•G pairs were repaired through the cell mismatch repair mechanism to convert the original C•G pair into T•A, or reduced to the original C•G through the uracil glycosylase mediated base excision repair. The presence of UGI minimizes the second result and increases the production of required T•A base pairs.
(addgene blog by Guest Blogger)