Difference between revisions of "Part:BBa K3595001"
Liyingying (Talk | contribs) |
Liyingying (Talk | contribs) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
=Usage and Biology= | =Usage and Biology= | ||
| − | This part can be used as a coding sequence after the promoter pTac and RBS B0034. The argA protein can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA using argA and argAfbr, respectively. The constructed plasmid was transformed into <i>E.coli DH10b</i> to test its ammonia degradation efficiency. | + | This part can be used as a coding sequence after the promoter pTac and RBS B0034. The argA protein can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA using argA and argAfbr, respectively. The constructed plasmid was transformed into <i>E.coli DH10b-ΔargR </i> host cell to test its ammonia degradation efficiency. |
| + | [[File:T--GZ_HFI--argA.png|600px|thumb|center|The structure of the plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ]] | ||
==Experimental Setup== | ==Experimental Setup== | ||
*Genetic design principle of argA fbr was described on the page of [[Part:BBa_K3595082]] | *Genetic design principle of argA fbr was described on the page of [[Part:BBa_K3595082]] | ||
| − | *Plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ^ fbr was transfered into the E.coli DH10b -ΔargR host cell,respestively. Meanwhile,plasmid pBR322-KanR-pTac-argA was transfered into the E.coli DH10b | + | *Plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ^ fbr was transfered into the E.coli DH10b -ΔargR host cell,respestively. Meanwhile,plasmid pBR322-KanR-pTac-argA was transfered into the<i>E.coli DH10b </i> as a negative control, and M9 media as a blank control |
*Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | *Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | ||
| − | * | + | *Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin , 1.5 uL 1M IPTG and 5mM ammonia. |
*Detecting ammonia concentration in culture medium | *Detecting ammonia concentration in culture medium | ||
| + | ==Results== | ||
| + | *The DH10b with plasmid pTYT-argAfbr and ∆argR showed the most significant increase in conversion of ammonia, 40.47% more conversion rate compared to the wildtype | ||
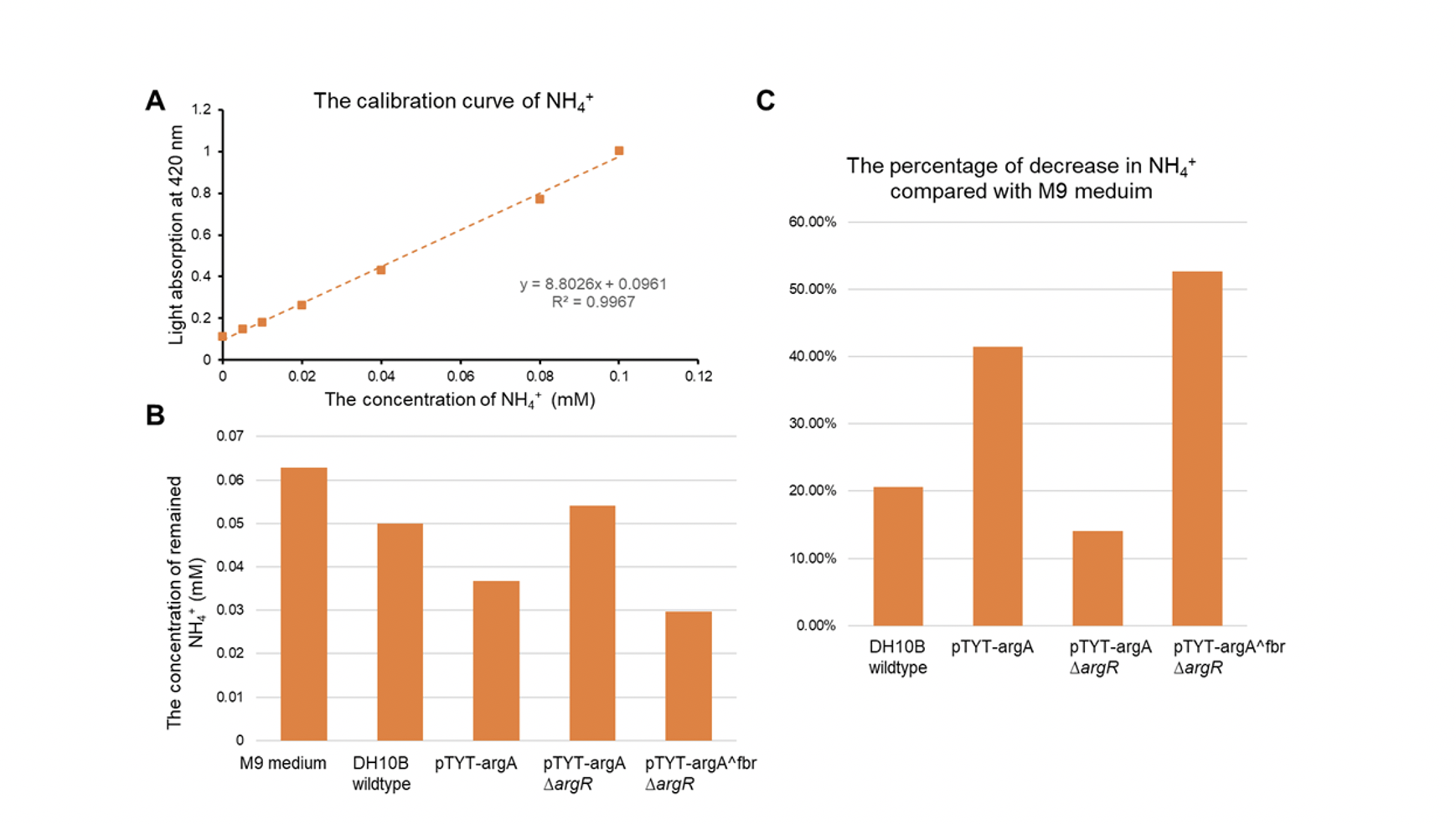
| + | [[File:T--GZ_HFI--NH3.png|600px|thumb|center|Detection results of the effect of the ammonia pathway. (A) The least-squared regression line of NH4+. (B)The concentration of NH4+. (C) The percentage of decrease in NH4+ compared with M9 medium. ]] | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K3595001 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3595001 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | <!-- Add more about the biology of this part here--> | ||
| + | =Reference= | ||
| + | Cunin R, et al. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 1986;50(3):314-52.<br> | ||
| + | Rajagopal BS, et al. Use of Inducible Feedback-Resistant N-Acetylglutamate Synthetase (argA) Genes for Enhanced Arginine Biosynthesis by Genetically Engineered Escherichia coli K-12 Strains. Appl Environ Microbiol 1998;64(5):1805-11.<br> | ||
| + | Vyas S, et al. Feedback inhibition of acetylglutamate synthetase by arginine in Escherichia coli. Arch Biochem Biophys 1963; 100(3), 542-6. | ||
Latest revision as of 19:11, 27 October 2020
argA-Wildtype
The ammonia that enters into E.coli is converted to glutamate along with α-Ketoglutarate . The series of reactions that convert glutamate into arginine are catalyzed by N-acetyl glutamate synthetase (NAGS), encoded by gene argA, to acetylize glutamate
Usage and Biology
This part can be used as a coding sequence after the promoter pTac and RBS B0034. The argA protein can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA using argA and argAfbr, respectively. The constructed plasmid was transformed into E.coli DH10b-ΔargR host cell to test its ammonia degradation efficiency.
Experimental Setup
- Genetic design principle of argA fbr was described on the page of Part:BBa_K3595082
- Plasmid pBR322-KanR-pTac-argA and pBR322-KanR-pTac-argA ^ fbr was transfered into the E.coli DH10b -ΔargR host cell,respestively. Meanwhile,plasmid pBR322-KanR-pTac-argA was transfered into theE.coli DH10b as a negative control, and M9 media as a blank control
- Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm.
- Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin , 1.5 uL 1M IPTG and 5mM ammonia.
- Detecting ammonia concentration in culture medium
Results
- The DH10b with plasmid pTYT-argAfbr and ∆argR showed the most significant increase in conversion of ammonia, 40.47% more conversion rate compared to the wildtype
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 626
Reference
Cunin R, et al. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev 1986;50(3):314-52.
Rajagopal BS, et al. Use of Inducible Feedback-Resistant N-Acetylglutamate Synthetase (argA) Genes for Enhanced Arginine Biosynthesis by Genetically Engineered Escherichia coli K-12 Strains. Appl Environ Microbiol 1998;64(5):1805-11.
Vyas S, et al. Feedback inhibition of acetylglutamate synthetase by arginine in Escherichia coli. Arch Biochem Biophys 1963; 100(3), 542-6.