Difference between revisions of "Part:BBa K3595010"
Liyingying (Talk | contribs) |
Liyingying (Talk | contribs) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 8: | Line 8: | ||
=Usage and Biology= | =Usage and Biology= | ||
This part can be used as a coding sequence after the promoter pTac and RBS B0034. The feedback inhibition-insensitive SAT can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-Mutant,among which the mutants include cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2. The constructed plasmid was transformed into <i>Nissle </i> host cell to test its production of cysteine. | This part can be used as a coding sequence after the promoter pTac and RBS B0034. The feedback inhibition-insensitive SAT can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-Mutant,among which the mutants include cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2. The constructed plasmid was transformed into <i>Nissle </i> host cell to test its production of cysteine. | ||
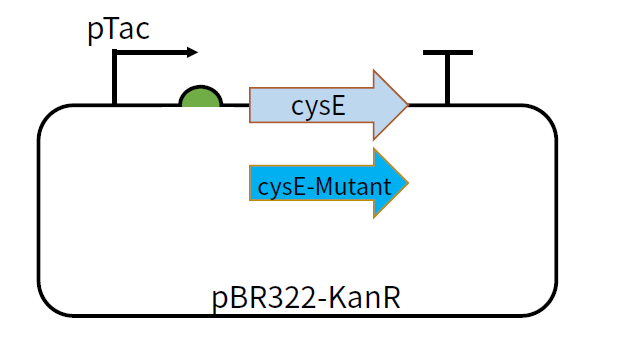
| − | [[File:T--GZ_HFI-- | + | [[File:T--GZ_HFI--cysE.png|600px|thumb|center|The structure of the plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant ]] |
==Experimental Setup== | ==Experimental Setup== | ||
*Genetic information of cysE,cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2 was described on the page of [[Part:BBa_K3595004]],[[Part:BBa_K3595005]],[[Part:BBa_K3595006]],[[Part:BBa_K3595007]], [[Part:BBa_K3595008]],[[Part:BBa_K3595009]], [[Part:BBa_K35950010]], [[Part:BBa_K3595011]],respectively. | *Genetic information of cysE,cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2 was described on the page of [[Part:BBa_K3595004]],[[Part:BBa_K3595005]],[[Part:BBa_K3595006]],[[Part:BBa_K3595007]], [[Part:BBa_K3595008]],[[Part:BBa_K3595009]], [[Part:BBa_K35950010]], [[Part:BBa_K3595011]],respectively. | ||
*Plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant was transfered into the <i>Nissle </i> host cell,respestively. | *Plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant was transfered into the <i>Nissle </i> host cell,respestively. | ||
*Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | *Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm. | ||
| − | *Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin | + | *Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin and 1.5 uL 1M IPTG. At the same time, wild-type Nissle was inoculated as negative control, and M9 medium was used as blank control. |
*Detecting cysteine concentration in culture medium | *Detecting cysteine concentration in culture medium | ||
==Results== | ==Results== | ||
*All of our engineered bacteria have shown great improvement in the ability to produce cysteine, which represents that they can absorb H2S to a larger extent. | *All of our engineered bacteria have shown great improvement in the ability to produce cysteine, which represents that they can absorb H2S to a larger extent. | ||
*EcN with plasmid pTYT-cysE-5-11-2 had the best effect, 98.72% batter than EcN wildtype and 8.79% better than overexpression of cysE (pTYT-cysE) | *EcN with plasmid pTYT-cysE-5-11-2 had the best effect, 98.72% batter than EcN wildtype and 8.79% better than overexpression of cysE (pTYT-cysE) | ||
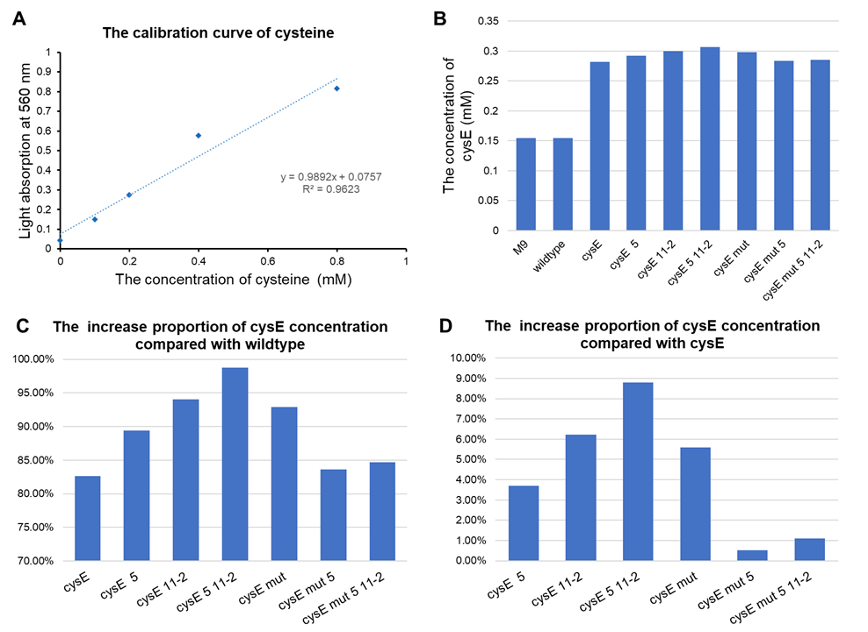
| − | + | [[File:T--GZ_HFI--H2S.png|600px|thumb|center|Detection results of the effect of the hydrogen sulfide pathway. (A) The calibration line of cysteine. (B) Concentration of cysteine detected in each group. (C) The ratio of increased cysteine concentration in each group compared to wildtype. (D) Cysteine production compared to pTYT-cysE. CysE-mut in the Figure stands for cysE-256. ]] | |
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K3595010 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3595010 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | <!-- Add more about the biology of this part here--> | ||
| + | =Reference= | ||
| + | Kai Y, et al. Engineering of Escherichia coli L-serine O-acetyltransferase on the basis of crystal structure: desensitization to feedback inhibition by L-cysteine. Protein Eng Des Sel 2006;19(4):163-7.<br> | ||
| + | Nakamori S, et al. Overproduction of L-Cysteine and L-Cystine by Escherichia coli Strains with a Genetically Altered Serine Acetyltransferase. Appl Environ Microbiol 1998; 64(5):1607-11. | ||
| + | |||
Latest revision as of 19:03, 27 October 2020
Mutant cysE-256-11-2
cysE-256-11-2 is a mutant of cysE gene, which is obtained by combining the positions of mutations in mutants cysE-256, and cysE-11-2.In the pathway of converting hydrogen sulfide to L-cysteine, L-serine and Acetyl-CoA form O-acetylserine catalyzed by L-serine O-acetyltransferase (SAT). SAT is encoded by gene cysE, and its feedback is inhibited by the final product L-cysteine . In order to induce overproduction of L-cysteine, we need to develop a mutant gene of cysE which will code for feedback inhibition-insensitive SAT.We tried to construct different mutants through site mutation, and cysE-256-11-2 is one of the different mutants.
Usage and Biology
This part can be used as a coding sequence after the promoter pTac and RBS B0034. The feedback inhibition-insensitive SAT can be translated under the induction of IPTG. We constructed plasmids pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-Mutant,among which the mutants include cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2. The constructed plasmid was transformed into Nissle host cell to test its production of cysteine.
Experimental Setup
- Genetic information of cysE,cysE-256, cysE-5, cysE-11-2, cysE-5-11-2, cysE-256-5,cysE-256-11-2,cysE-256-5-11-2 was described on the page of Part:BBa_K3595004,Part:BBa_K3595005,Part:BBa_K3595006,Part:BBa_K3595007, Part:BBa_K3595008,Part:BBa_K3595009, Part:BBa_K35950010, Part:BBa_K3595011,respectively.
- Plasmid pBR322-KanR-pTac-cysE and pBR322-KanR-pTac-cysE-mutant was transfered into the Nissle host cell,respestively.
- Single colonies were selected from the experimental LB-agar plate , then inoculated into test-tube tubes with 4000 μL LB liquid medium with 4uL kanamycin for overnight growth at 37 °C and 200 rpm.
- Inoculating 15 uL of culture solution overnight into a 24-well plate containing 3 mL M9 medium for overnight growth at 37 °C and 200 rpm.. The media contained 3 ul kanamycin and 1.5 uL 1M IPTG. At the same time, wild-type Nissle was inoculated as negative control, and M9 medium was used as blank control.
- Detecting cysteine concentration in culture medium
Results
- All of our engineered bacteria have shown great improvement in the ability to produce cysteine, which represents that they can absorb H2S to a larger extent.
- EcN with plasmid pTYT-cysE-5-11-2 had the best effect, 98.72% batter than EcN wildtype and 8.79% better than overexpression of cysE (pTYT-cysE)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 684
- 1000COMPATIBLE WITH RFC[1000]
Reference
Kai Y, et al. Engineering of Escherichia coli L-serine O-acetyltransferase on the basis of crystal structure: desensitization to feedback inhibition by L-cysteine. Protein Eng Des Sel 2006;19(4):163-7.
Nakamori S, et al. Overproduction of L-Cysteine and L-Cystine by Escherichia coli Strains with a Genetically Altered Serine Acetyltransferase. Appl Environ Microbiol 1998; 64(5):1607-11.