Difference between revisions of "Part:BBa K863030"
(→Biology and Usage) |
|||
| (10 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K863030 short</partinfo> | <partinfo>BBa_K863030 short</partinfo> | ||
cDNA sequence from tvl5 (Trametes versicolor) | cDNA sequence from tvl5 (Trametes versicolor) | ||
| + | |||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 19: | ||
<partinfo>BBa_K863030 parameters</partinfo> | <partinfo>BBa_K863030 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | ===Improvement Made by Shanghai_United 2020=== | ||
| + | Compared with the old part BBa_K863030, expressing laccase (EC1.10.3.2), we design a new part BBa_K3584000, expressing polyphenol oxidase (EC 1.14.18.1, PPO). Both of the two enzymes can catalyze phenol or polyphenol to the corresponding quinone. | ||
| + | The team iGEM12_Bielefeld-Germany aimed to use laccase to degrade polycyclic aromatic hydrocarbons for filtering populated water. They successfully identified one of the laccases, part BBa_K863030. They purified and detected the activity of this laccase protein. | ||
| + | |||
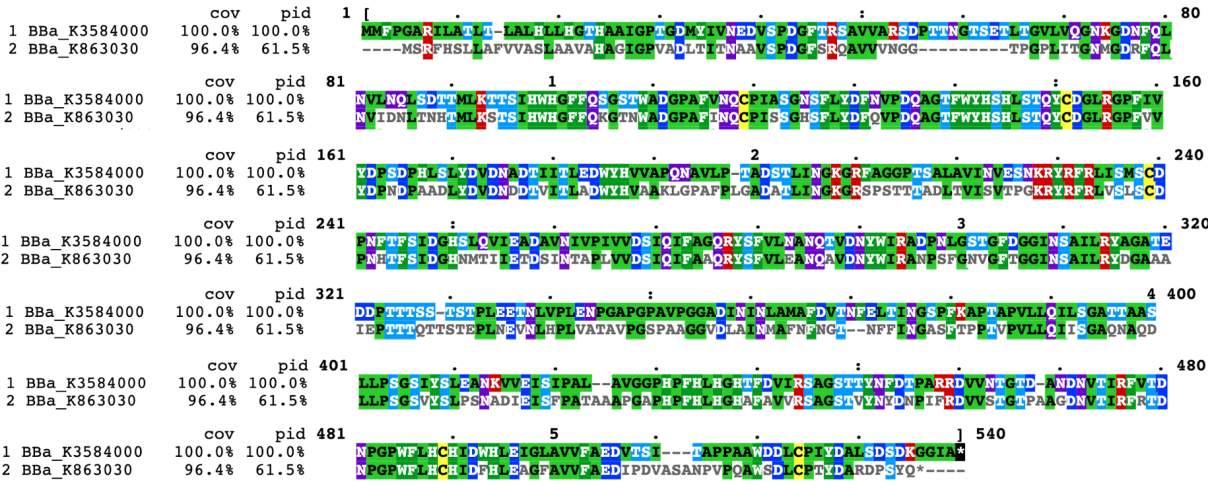
| + | In contrast, our team aimed to degrade p-cresol sulfate, on the basis of which we hope to provide chronic kidney disease patients with potential health supplements or even treatment. We chose PPO, whose sequence is different from laccase(Fig1), to construct our desired engineering strain. | ||
| + | |||
| + | [[File:T--Shanghai_United-BBa_K3584000 improve fig 1.png|600px|thumb|center|Figure 1 The blast results about the protein sequence of our new part BBa_K3584000 and the old one BBa_K863030.]] | ||
| + | |||
| + | |||
| + | Initially, we constructed a composite part BBa_K3584002 and transformed it into E.coli BL21(DE3). We successfully expressed and purified PPO protein, and detected its enzyme activity. | ||
| + | |||
| + | Furthermore, in order to optimize the function of BBa_K3584000, we constructed an improved composite part BBa_K3584008, which was designed as tyrosine-induced regulation system. This system allowed us to control the expression of PPO by adding or removing tyrosine. | ||
| + | |||
| + | In addition, we select a probiotics E.coli Nissle 1917 as the host. Therefore, our engineering strain would be a competitive health supplement or drug candidate for degrading p-cresol and hence bring about benefits to chronic kidney patients. | ||
| + | |||
| + | ===Biology and Usage=== | ||
| + | BBa_K3584000 is the coding sequence of a polyphenol oxidase (PPO) from Pleurotus ostreatus, which is different from the present enzymes showing the similar activity. PPO is a group of enzyme that are able to oxidize phenol or polyphenol by molecular oxygen to form the corresponding quinone. In a broad sense, polyphenol oxidase can be divided into three categories: monophenol monooxidase (tyrosinase, EC.1.14.18.1), bisphenol oxidase (catechol oxidase, EC. 1.10.3.2) and laccase (laccase, EC.1.10.3.1). Among these three types of polyphenol oxidase, catechol oxidase is mainly distributed in plants, and the polyphenol oxidase in microorganisms mainly includes laccase and tyrosinase. The polyphenol oxidase mentioned in most of the literature is generally the collective name of catechol oxidase and laccase. | ||
| + | [[File:T--Shanghai United--BBa K35284000 fig 0.jpg|500px|thumb|center|]] | ||
| + | |||
| + | ===Engineering Success=== | ||
| + | Production, purification, and SDS-PAGE analysis of recombinant PPO | ||
| + | |||
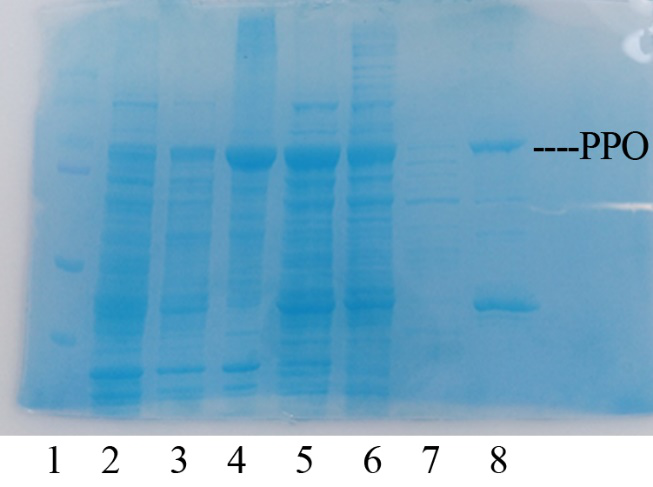
| + | In order to present the function of the part, the PPO gene was expressed in E. coli BL21 (DE3) under the control of T7 promoter. Then the bacterial cells are collected and crushed, and the PPO enzyme solution is purified by further confirmation by the SDS-PAGE method, which is found in the corresponding protein band of approximately 57 kDa (Figure 1). | ||
| + | [[File:T--Shanghai United--BBa K35284000 fig 1.jpg|500px|thumb|center|Figure 2]] | ||
| + | Figure 2. Production, purification, and SDS-PAGE analysis of recombinant PPO. Lane: protein molecular weight standard; lane 8: purified PPO. | ||
| + | |||
| + | Enzymatic activity of the PPO | ||
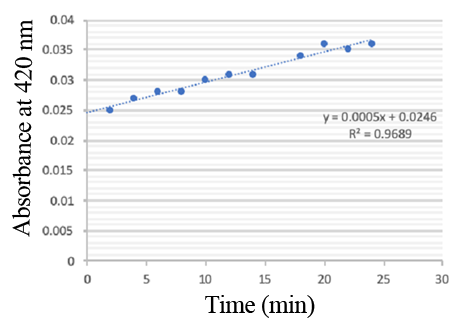
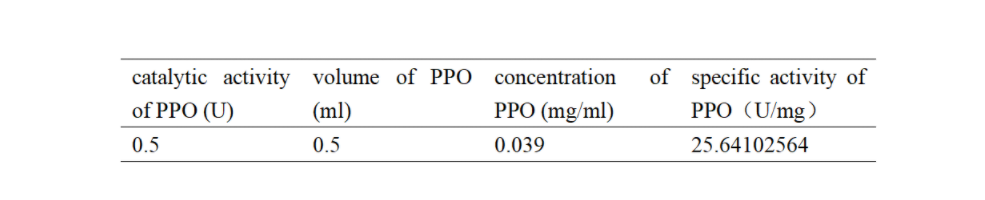
| + | PPO activity was determined using an analogue of p-cresol (catechol) as a substrate. In proper reaction mixture, PPO and 10 mM catechol was incubated at 50 ℃, pH 7.0 for 3 min, and the change in absorbance at 420 nm was measured spectrophotometrically. One unit of enzyme (PPO) activity was defined as the amount of enzyme that increased absorbance of 0.001 per minute. We found that as the extension of reaction time, reaction liquid of absorbance at 420 nm increased linear (Figure 3), we obtained by catalytic activity of PPO enzyme fluid, and divided by the corresponding protein concentration, the specific activity of PPO (Table 1). | ||
| + | [[File:T--Shanghai United--BBa K35284000 fig 2.jpg|500px|thumb|center|Figure 3]] | ||
| + | PPO activity was determined using catechol as a substrate. The reaction mixture contained PPO protein and 10 mM catechol which was incubated at 50 ℃, pH 7.0 for 3 min, and the change in absorbance at 420 nm was recorded. | ||
| + | |||
| + | Table 1. Specific activity of recombinant PPO | ||
| + | [[File:T--Shanghai United--BBa K35284006 fig 4.jpg|500px|thumb|center|Table 1]] | ||
| + | |||
| + | == TVEL5 integrated in shuttle vector == | ||
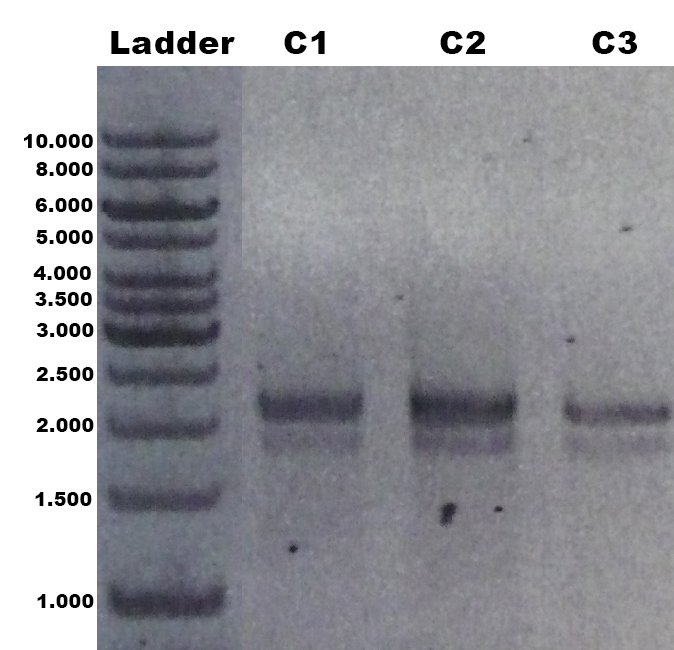
| + | The laccase ''Trametes versicolor'' TVEL5 [https://parts.igem.org/wiki/index.php?title=Part:BBa_K863030 BBa_K863030] was cloned via the ''Aar''I restriction site into the shuttle vector ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K863204 BBa_K863204]). This construct ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K863207 BBa_K863207]) was linearized and integrated into the yeast genome of ''P. pastoris'' GS115. Positive clones with single cross overs were identified by PCR on genomic DNA with the primer 5AOX-Phenotype-FW (GACTGGTTCCAATTGACAAGC) and TT-Phenotype-RV (GCAAATGGCATTCTGACATCC) as shown in Figure 1 and 2. Positive clones with single cross over resulted in a 2.2 kb DNA band, the ''aox1'' gene, and a 1.9 kb DNA band, the TVEL5 laccase DNA band. As control the shuttle vector ([https://parts.igem.org/wiki/index.php?title=Part:BBa_K863204 BBa_K863204]) itself was used. The expected DNA size is 437 bp. | ||
| + | [[Image:Bielefeld2012_TV5_negKon_pSH19A_Agarosegel.jpg|thumb|left|220px|'''Figure 1:''' [https://parts.igem.org/wiki/index.php?title=Part:BBa_K863204 BBa_K863204] was used as a control for PCR with the primer 5AOX-Phenotype-FW and TT-Phenotype-RV. Expected DNA band: 437 bp.]] | ||
| + | [[Image:Bielefeld2012_TV5_Probe_Agarosegel.jpg|thumb|right|300px|'''Figure 2:''' PCR product of the gDNA from the culture 1, 2 and 3 (C1, C2, C3) with the primer 5AOX-Phenotype-FW and TT-Phenotype-RV. Expected DNA bands: 1.9 kb (''tvel5'') and 2.2 kb (''aox1'').]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | == Cultivation and protein induction == | ||
| + | The clones were cultivated in minimal medium and induced with 1% (v/v) methanol twice a day for 2 days. | ||
| + | |||
| + | == Enzyme activity test == | ||
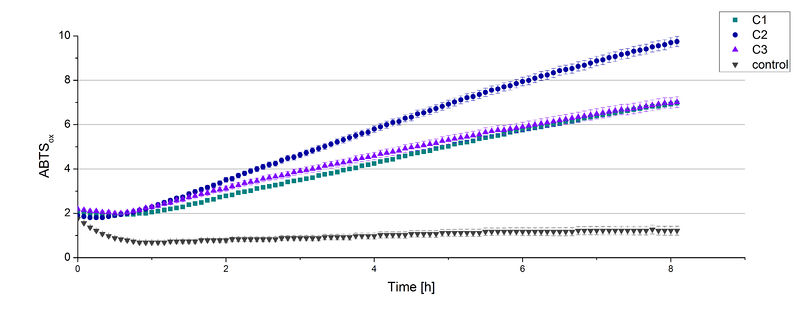
| + | The supernatant of the culture was examined to active laccases by testing the enzyme activity. For this purpose the supernatant was re-buffered into H<sub>2</sub>O with HiTrap Desalting Columns and incubated with 0.4 mM CuCl<sub>2</sub>. After 2 hours of incubation time 140 µL of the re-buffered supernatant were applied for the activity assay with 40 µL Britton Robinson buffer (pH 5) and measured at 25 °C. These setting were chosen because of the experiences with the TVEL0 laccase. To detect activity of the TVEL5 laccase 9 mM ABTS were used. Figure 3 shows a distinct activity of each cultivation (C1, C2, C3) in comparison to the control, which was treated like the other samples. The control, which was also treated with CuCl<sub>2</sub> also represented the negative control to determine the effect of CuCl<sub>2</sub> oxidizing ABTS. In Figure 4 the same experimental setup was used in a total volume of 1 mL. The oxidation of ABTS by TVEL5 laccase is visible to the naked eye in C1, C2 and C3 in comparison to the control. C2 shows the highest potential in oxidizing ABTS in the activity measurements and also in the experiments with a higher volume. This may be due to a higher TVEL5 concentration in the supernatant of this cultivation. | ||
| + | |||
| + | [[Image:Hefeakti.jpg|thumb|left|420px|'''Figure 3:'''Activity assay of TVEL5 using the re-buffered, CuCl<sub>2</sub> incubated supernatant of C1, C2 and C3 with 140 µL each. 40 µL Britton Robinson buffer (pH 5) and 9 mM ABTS were added. Oxidized ABTS was detected by measuring the change in OD at 420 nm at 25 °C for 8 hours.]] | ||
| + | [[Image:Bielefeld2012_TVEL5_Eppis.jpg|thumb|right|350px|'''Figure 4:'''Activity test of cultivation (culture 1, 2, 3) supernatant of [https://parts.igem.org/Part:BBa_K863207 BBa_K863207]. The control is the supernatant of ''P. pastoris'' GS115. The supernatants were re-buffered and CuCl<sub>2</sub> incubated. Britton Robinson buffer and 9 mM ABTS were applied. Picture was taken after 8 hours incubation at 25 °C.]] | ||
Latest revision as of 13:44, 27 October 2020
tvel5 laccase cDNA sequence from Trametes versicolor
cDNA sequence from tvl5 (Trametes versicolor)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 773
Illegal BglII site found at 1205
Illegal XhoI site found at 805
Illegal XhoI site found at 1438 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1291
Illegal NgoMIV site found at 1444
Illegal AgeI site found at 172 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 727
Illegal SapI.rc site found at 237
Improvement Made by Shanghai_United 2020
Compared with the old part BBa_K863030, expressing laccase (EC1.10.3.2), we design a new part BBa_K3584000, expressing polyphenol oxidase (EC 1.14.18.1, PPO). Both of the two enzymes can catalyze phenol or polyphenol to the corresponding quinone. The team iGEM12_Bielefeld-Germany aimed to use laccase to degrade polycyclic aromatic hydrocarbons for filtering populated water. They successfully identified one of the laccases, part BBa_K863030. They purified and detected the activity of this laccase protein.
In contrast, our team aimed to degrade p-cresol sulfate, on the basis of which we hope to provide chronic kidney disease patients with potential health supplements or even treatment. We chose PPO, whose sequence is different from laccase(Fig1), to construct our desired engineering strain.
Initially, we constructed a composite part BBa_K3584002 and transformed it into E.coli BL21(DE3). We successfully expressed and purified PPO protein, and detected its enzyme activity.
Furthermore, in order to optimize the function of BBa_K3584000, we constructed an improved composite part BBa_K3584008, which was designed as tyrosine-induced regulation system. This system allowed us to control the expression of PPO by adding or removing tyrosine.
In addition, we select a probiotics E.coli Nissle 1917 as the host. Therefore, our engineering strain would be a competitive health supplement or drug candidate for degrading p-cresol and hence bring about benefits to chronic kidney patients.
Biology and Usage
BBa_K3584000 is the coding sequence of a polyphenol oxidase (PPO) from Pleurotus ostreatus, which is different from the present enzymes showing the similar activity. PPO is a group of enzyme that are able to oxidize phenol or polyphenol by molecular oxygen to form the corresponding quinone. In a broad sense, polyphenol oxidase can be divided into three categories: monophenol monooxidase (tyrosinase, EC.1.14.18.1), bisphenol oxidase (catechol oxidase, EC. 1.10.3.2) and laccase (laccase, EC.1.10.3.1). Among these three types of polyphenol oxidase, catechol oxidase is mainly distributed in plants, and the polyphenol oxidase in microorganisms mainly includes laccase and tyrosinase. The polyphenol oxidase mentioned in most of the literature is generally the collective name of catechol oxidase and laccase.
Engineering Success
Production, purification, and SDS-PAGE analysis of recombinant PPO
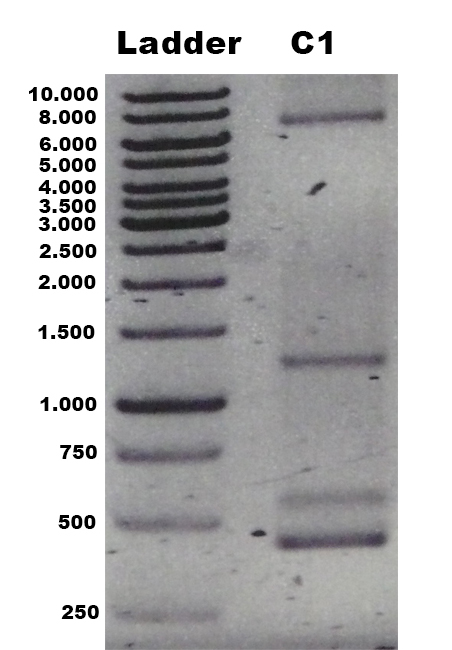
In order to present the function of the part, the PPO gene was expressed in E. coli BL21 (DE3) under the control of T7 promoter. Then the bacterial cells are collected and crushed, and the PPO enzyme solution is purified by further confirmation by the SDS-PAGE method, which is found in the corresponding protein band of approximately 57 kDa (Figure 1).
Figure 2. Production, purification, and SDS-PAGE analysis of recombinant PPO. Lane: protein molecular weight standard; lane 8: purified PPO.
Enzymatic activity of the PPO PPO activity was determined using an analogue of p-cresol (catechol) as a substrate. In proper reaction mixture, PPO and 10 mM catechol was incubated at 50 ℃, pH 7.0 for 3 min, and the change in absorbance at 420 nm was measured spectrophotometrically. One unit of enzyme (PPO) activity was defined as the amount of enzyme that increased absorbance of 0.001 per minute. We found that as the extension of reaction time, reaction liquid of absorbance at 420 nm increased linear (Figure 3), we obtained by catalytic activity of PPO enzyme fluid, and divided by the corresponding protein concentration, the specific activity of PPO (Table 1).
PPO activity was determined using catechol as a substrate. The reaction mixture contained PPO protein and 10 mM catechol which was incubated at 50 ℃, pH 7.0 for 3 min, and the change in absorbance at 420 nm was recorded.
Table 1. Specific activity of recombinant PPO
TVEL5 integrated in shuttle vector
The laccase Trametes versicolor TVEL5 BBa_K863030 was cloned via the AarI restriction site into the shuttle vector (BBa_K863204). This construct (BBa_K863207) was linearized and integrated into the yeast genome of P. pastoris GS115. Positive clones with single cross overs were identified by PCR on genomic DNA with the primer 5AOX-Phenotype-FW (GACTGGTTCCAATTGACAAGC) and TT-Phenotype-RV (GCAAATGGCATTCTGACATCC) as shown in Figure 1 and 2. Positive clones with single cross over resulted in a 2.2 kb DNA band, the aox1 gene, and a 1.9 kb DNA band, the TVEL5 laccase DNA band. As control the shuttle vector (BBa_K863204) itself was used. The expected DNA size is 437 bp.
Cultivation and protein induction
The clones were cultivated in minimal medium and induced with 1% (v/v) methanol twice a day for 2 days.
Enzyme activity test
The supernatant of the culture was examined to active laccases by testing the enzyme activity. For this purpose the supernatant was re-buffered into H2O with HiTrap Desalting Columns and incubated with 0.4 mM CuCl2. After 2 hours of incubation time 140 µL of the re-buffered supernatant were applied for the activity assay with 40 µL Britton Robinson buffer (pH 5) and measured at 25 °C. These setting were chosen because of the experiences with the TVEL0 laccase. To detect activity of the TVEL5 laccase 9 mM ABTS were used. Figure 3 shows a distinct activity of each cultivation (C1, C2, C3) in comparison to the control, which was treated like the other samples. The control, which was also treated with CuCl2 also represented the negative control to determine the effect of CuCl2 oxidizing ABTS. In Figure 4 the same experimental setup was used in a total volume of 1 mL. The oxidation of ABTS by TVEL5 laccase is visible to the naked eye in C1, C2 and C3 in comparison to the control. C2 shows the highest potential in oxidizing ABTS in the activity measurements and also in the experiments with a higher volume. This may be due to a higher TVEL5 concentration in the supernatant of this cultivation.