Difference between revisions of "Part:BBa K1529321"
| (3 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1529321 short</partinfo> | <partinfo>BBa_K1529321 short</partinfo> | ||
| − | Prhl(RR) is a promoter that is activated by C4HSL.<br> | + | This part is Prhl(RR) promoter regulating GFP. |
| − | We improved Prhl (<partinfo>BBa_R0071</partinfo>) by changing LuxR binding site of Plux (<partinfo>BBa_R0062</partinfo>) to RhlR binding site. (Fig.1)<br> | + | Prhl(RR) is a promoter that is activated by C4HSL-RhlR complex. <br> |
| − | The bottom structures of LuxR and RhlR are similar to each other, so RhlR can bind to Lux Box. (Fig.2) <br> | + | We improved Prhl (<partinfo>BBa_R0071</partinfo>) by changing a LuxR binding site of Plux (<partinfo>BBa_R0062</partinfo>) to a RhlR binding site. (Fig. 1.)<br> |
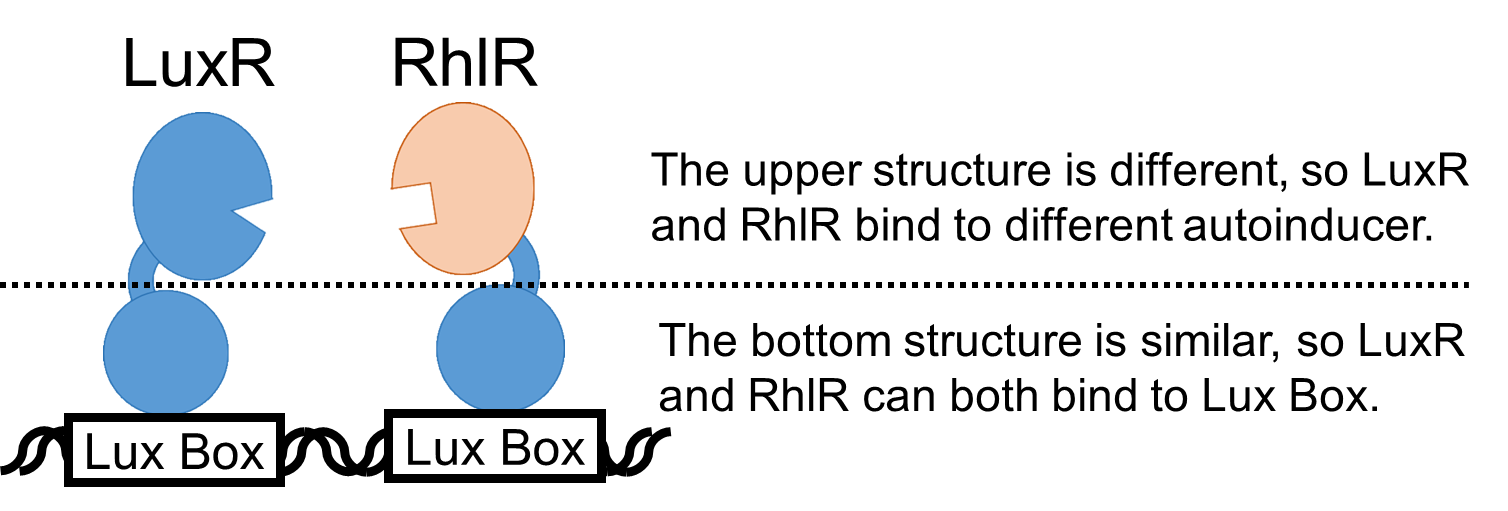
| + | The bottom structures of LuxR and RhlR are similar to each other, so RhlR can also bind to Lux Box. (Fig. 2.) <br> | ||
| − | + | We constructed this part to characterize the function of the Prhl(RR) promoter (<partinfo>BBa_K1529310</partinfo>), by inserting Prhl(RR) promoter upstream of a GFP coding sequence. <br> | |
[[Image:titech2014_parts_rhl_promoter_main_Fig1.png|thumb|center|550px|<b>Fig. 1.</b> Our newly designed promoter]] | [[Image:titech2014_parts_rhl_promoter_main_Fig1.png|thumb|center|550px|<b>Fig. 1.</b> Our newly designed promoter]] | ||
[[Image:titech2014_parts_luxR_and_rhlR_difference_main_Fig2.png|thumb|center|550px|<b>Fig. 2.</b> Difference between LuxR and RhlR]] | [[Image:titech2014_parts_luxR_and_rhlR_difference_main_Fig2.png|thumb|center|550px|<b>Fig. 2.</b> Difference between LuxR and RhlR]] | ||
| − | |||
| − | |||
| − | |||
| − | [[Image: | + | By using reporter cells that contain Prhl(RR)-GFP, we measured the fluorescence intensity of the cells induced by C4HSL. <br> |
| + | Through an induction assay, we found that our new Prhl(RL) promoter was actually activated by C4HSL-RhlR complex and had high induced/not-induced ratio(Fig. 3.). <br> | ||
| + | This means Prhl(RL) promoter has the higher expression level while keeping the low leakage. <br> | ||
| + | |||
| + | We can say that Prhl(RL) promoter is the best improved Prhl promoter due to the advantages of less leak and higher expression level. <br> | ||
| + | |||
| + | |||
| + | [[Image:Improved_Prhl(RR)_Promoter_Assay_Result.png|thumb|center|600px|<b>Fig. 3.</b> Fluorescence intensity detected by flow cytometer]] | ||
| + | |||
<br> | <br> | ||
| − | For more information, see [http:// | + | For more information, see [http://2014.igem.org/Team:Tokyo_Tech/Experiment/Prhl_reporter_assay our work in Tokyo_Tech 2014 wiki]. |
| + | |||
| + | ===Contribution: Waseda 2020=== | ||
| + | For the crosstalk experiments, we constructed two kinds of cell-free translation systems containing substantial amount of R proteins (LuxR,RhlR). In those systems, we put signaling molecules (3OHSL-C4 or 3OHSL-C6) and target genes (Plux/tet-GFP or Prhl-RR-GFP), respectively. We measured the fluorescence of GFP expressed from plux/tet-GFP and prhl(RR)-GFP on a RT-qPCR machine. | ||
| + | |||
| + | Fig 1 shows the result of the crosstalk test in a cell-free system containing sufficient amount of LuxR protein. In the results, non-cognate Prhl(RR) promter was highly activated by luxR-3OC6HSL complex as well as the cognate Plux/tet promoter. | ||
| + | Fig 2 shows the result of the crosstalk test in a cell-free system containing sufficient amount of RhlR protein. As a result, we revealed that Prhl(RR)-GFP can work in vitro. | ||
| + | |||
| + | [[Image:T--Waseda--crosstalk_luxR.png|thumb|center|600px|<b>Fig. 1. </b>Crosstalk assay in a cell-free system containing LuxR protein. | ||
| + | ]]<br> | ||
| + | |||
| + | [[Image:T--Waseda--crosstalk_rhlR.png|thumb|center|600px|<b>Fig. 2. </b>Crosstalk assay in a cell-free system containing rhlR protein. | ||
| + | ]]<br> | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 08:33, 27 October 2020
Prhl(RR)_GFP
This part is Prhl(RR) promoter regulating GFP.
Prhl(RR) is a promoter that is activated by C4HSL-RhlR complex.
We improved Prhl (BBa_R0071) by changing a LuxR binding site of Plux (BBa_R0062) to a RhlR binding site. (Fig. 1.)
The bottom structures of LuxR and RhlR are similar to each other, so RhlR can also bind to Lux Box. (Fig. 2.)
We constructed this part to characterize the function of the Prhl(RR) promoter (BBa_K1529310), by inserting Prhl(RR) promoter upstream of a GFP coding sequence.
By using reporter cells that contain Prhl(RR)-GFP, we measured the fluorescence intensity of the cells induced by C4HSL.
Through an induction assay, we found that our new Prhl(RL) promoter was actually activated by C4HSL-RhlR complex and had high induced/not-induced ratio(Fig. 3.).
This means Prhl(RL) promoter has the higher expression level while keeping the low leakage.
We can say that Prhl(RL) promoter is the best improved Prhl promoter due to the advantages of less leak and higher expression level.
For more information, see [http://2014.igem.org/Team:Tokyo_Tech/Experiment/Prhl_reporter_assay our work in Tokyo_Tech 2014 wiki].
Contribution: Waseda 2020
For the crosstalk experiments, we constructed two kinds of cell-free translation systems containing substantial amount of R proteins (LuxR,RhlR). In those systems, we put signaling molecules (3OHSL-C4 or 3OHSL-C6) and target genes (Plux/tet-GFP or Prhl-RR-GFP), respectively. We measured the fluorescence of GFP expressed from plux/tet-GFP and prhl(RR)-GFP on a RT-qPCR machine.
Fig 1 shows the result of the crosstalk test in a cell-free system containing sufficient amount of LuxR protein. In the results, non-cognate Prhl(RR) promter was highly activated by luxR-3OC6HSL complex as well as the cognate Plux/tet promoter. Fig 2 shows the result of the crosstalk test in a cell-free system containing sufficient amount of RhlR protein. As a result, we revealed that Prhl(RR)-GFP can work in vitro.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 90
Illegal BamHI site found at 78 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 777