Difference between revisions of "Part:BBa K2753015"
Cassandrashi (Talk | contribs) |
Cassandrashi (Talk | contribs) |
||
| Line 93: | Line 93: | ||
--------------------- | --------------------- | ||
| − | + | <span style="font-size: 130%;font-weight:bold">Part improvement</span> | |
<p>Group: KEYSTONE 2020</p> | <p>Group: KEYSTONE 2020</p> | ||
<p>Summary: Used Gibson Assembly to substitute GES with bLIS (linalool synthase) to achieve linalool production. </p> | <p>Summary: Used Gibson Assembly to substitute GES with bLIS (linalool synthase) to achieve linalool production. </p> | ||
Latest revision as of 08:21, 27 October 2020
pSB1C3-RiboJ-rbs-GPPS-rbs-ObGES
This part consists of a ribozyme (BBa_K1679038), two RBSs, an Abies Grandis geranyl pyrophosphate synthase coding sequence (BBa_K2753002) and an Ocimum basilicum geraniol synthase (BBa_2753003)It metabolizes IPP and DMAPP which are five-carbon blocks produced by either MVA pathway or MEP pathway or in some case both MVA and MEP pathway. IPP and DMAPP are converted to geraniol, a monoterpenoid with a rose-like odour and is a prosperous antimicrobial reagent and gasoline alternative1. With addition of more downstream enzymes, geraniol can be changed to a large range of monoterpenes like linalool, actinide and nepetalactol.
Characterization

Figure. 1 The biosynthetic route for obtaining geraniol from acetyl-coA, which requires either a MVA or MEP pathway, a GPPS, and a GES.
In our study, we aim to achieve geraniol synthesis in E. coli, so at the beginning, we expressed a geraniol synthesis operon (https://parts.igem.org/Parts:BBa_K2753016) containing an Abies Grandis geranyl pyrophosphate synthase (https://parts.igem.org/Parts:BBa_K2753002) and an Ocimum basilicum geraniol synthase (https://parts.igem.org/Parts:BBa_K2753003) regulated by a pTac promoter in E. coli. Assuming that higher expression leads to higher yield, the operon was assembled on a high copy vector pUC20 whose copy number is approximately 500 per cell. However, when carrying out shake-flask fermentation with this strain, after induced by 25uM IPTG for 24 hours2, no geraniol production was detected using gas chromatography. (see detailed methods in our notebook: http://2018.igem.org/Team:GreatBay_China/Notebook)
We infer that the failure to produce geraniol was due to the lack of substrates IPP and DMAPP. Thus we decided to co-express an MVA pathway in addition to the geraniol synthesis operon. The pathway can either be added by cotransformation of a plasmid containing only the MVA pathway and the other plasmid with only GPPS and GES, or by constructing an all-in-one plasmid containing the MVA pathway, GPPS, and GES. We obtained a plasmid pMVA-only which was a gift from Taek Soon Lee, and have added GPPS and GES onto pMVA-only to acquire a pMVA-GPPS-GES.

Figure. 2 Geraniol production analysis by gas chromatography. (A) GPPS and GES are arranged in operon regulated by pTac, placed on a high copy vector pUC20. (B) The MVA pathway is split into two clusters and placed on a low copy vector with the upper cluster containing three genes and the downstream containing four. (C) A combined plasmid with GPPS&GES operon and MVA pathway with a low copy p15A origin. (D) Gas chromatography for geraniol produced by E. coli expressing pMVA only (middle trace) or pMVA-GPPS-GES (bottom trace) whose peaks coincide with a geraniol standard (top trace). (E) Geraniol yield is noticeable with only the heterologous MVA pathway. Upon introduction of GPPS and GES, the yield doubled relative to the negative control pMVA only.
Under the same fermentation condition, pMVA-GPPS-GES gave a geraniol titer of 11.4mg/L while the negative control pMVA-only as well produced geraniol titer of about 5.65mg/L. Then we are assured that a stable supply of precursors IPP and DMAPP are crucial for geraniol production. But still, there is room for higher geraniol yield. Therefore, to gain maximum flux to geraniol, we tuned the expression level of GPPS and GES using three promoters:
- TALE sp1 (https://parts.igem.org/Part:BBa_K2753030), copy number-independent promoter of medium strength
- TALE sp2 (https://parts.igem.org/Part:BBa_K2753018), copy number-independent promoter of low strength
- pTac (https://parts.igem.org/Part:BBa_K180000), IPTG inducible promoter
And three vectors of unique copy number:
- pUC20: ~500
- pR6K: ~15
- pSC101: ~1

Figure. 3 Geraniol production assay of TALEsp relative to pTac IPTG-inducible promoter. (A) Twelve combinations of promoters and plasmid backbones used to express a geraniol synthesis operon for characterization. pSC101 is the low copy vector, pR6K is the medium copy vector and pUC20 is the high copy vector. The combinations are transformed and tested in E. coli already harboring pMVA plasmids. (B) Geraniol yield achieved under the regulation of pTac, TALE1 sp1, TALE2 sp1, and TALE2 sp6 across different copy numbers is shown. The vertical error bars represents standard deviations of geraniol yield measured, and the horizontal error bars signify the reported range of copy number of plasmid backbones used. The portion of graph belong to TALE2sp6 is not shown as there is no detectable geraniol production.
Having transferred the constructs above along with pMVA-only into strain E. coli DH5α, the same shake-flask fermentation experiment was conducted. The result shows detectable geraniol production by each strain.
With pTac promoters, geraniol yield increased with the copy number of the vector, showing positively correlated relation. However, when TALE stabilized promoter (TALEsp) was used, higher the copy number of the vector, lower the production of geraniol, being the opposite of pTac. And the stronger TALEsp promoter, TALE1sp1 gave generally reduced yield compared to its weaker counterpart TALE2sp1. Interestingly, the graphs of the two TALEsp promoters appeared to be seemingly parallel to each other.

Figure. 4 Gas chromatography analysis for geraniol produced regulated by pTac and TALE2sp1 compared with the peak of geraniol standard. (A) Geraniol production accomplished by pTac regulation across low (pSC101), medium (pR6K) and high (pUC20) copy number. The area of the peak which appears at around 4.41s concurring with that of the geraniol standard increases along the copy number. (B) Geraniol yield assay using TALE2sp1 with the same set of copy number. The largest peak of geraniol emerges on the low copy vector pSC101. The peak shrinks in size on higher copy.
We surmised that the yield of geraniol was affected by two factors: the expression level of enzymes and the cellular burden. As for enzyme expression, there exists an optimal gene expression level that produces just enough enzyme to metabolize all the substrate. The yield would be the greatest at this level. But if lower than this level, production would increase with enzyme expression since there is a surplus of substrates. And if the enzyme expression is higher, the more enzymes now becomes a cellular burden as it no longer contributes to more product.
In the case of pTac, it fits into the scenario when the gene expression is lower than the optimum: higher copy produced more enzymes to catalyze geraniol synthesis reaction. As for TALE stabilized promoters, the expression level of enzymes would remain unchanged regardless of copy number due to the stabilization nature of TALE (More about TALE:[http://2018.igem.org/Team:GreatBay_China/Part_Collection]). So in low copy vector where the cellular burden isn’t significant, the product yield is only related to the strength of the promoter, explaining why TALE sp2 has better performance than TALE sp1 as it’s closer to the optimum. But when the copy number increases, the expression level is unaffected while the cellular burden rises sharply because much more TALE would be made to stabilize expression, leaving less energy available for geraniol synthesis. And it in turns answers why a negative trend of production is shown when regulated by TALE.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Reference
[1]: Zhou, J., Wang, C., Yoon, S., Jang, H., Choi, E., & Kim, S. (2014). Engineering Escherichia coli for selective geraniol production with minimized endogenous dehydrogenation. Journal of Biotechnology, 169, 42–50. https://doi.org/10.1016/j.jbiotec.2013.11.009
[2]: Alonso-gutierrez, J., Chan, R., Batth, T. S., Adams, P. D., Keasling, J. D., Petzold, C. J., & Soon, T. (2013). Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic Engineering, 19, 33–41. https://doi.org/10.1016/j.ymben.2013.05.004
Part improvement
Group: KEYSTONE 2020
Summary: Used Gibson Assembly to substitute GES with bLIS (linalool synthase) to achieve linalool production.
We applied the pathway to a new fragrance synthase. We aimed to produce linalool, a monoterpene that is commonly used as a scent and flavoring agent. The linalool synthase used in our experiments is the linalool synthase from Streptromyces clavuligerus that is called bLiS. It is a very effective linalool synthase. The synthesis of linalool can also be increased by the optimization of the fusion tags and the ribosome binding sites of the protein [1]. We applied the Gibson Assembly method to substitute the GES gene with bLIS ( (3R)-linalool synthase) from Streptomyces clavuligerus [2] and was able to synthesize linalool from geranyl pyrophosphate.
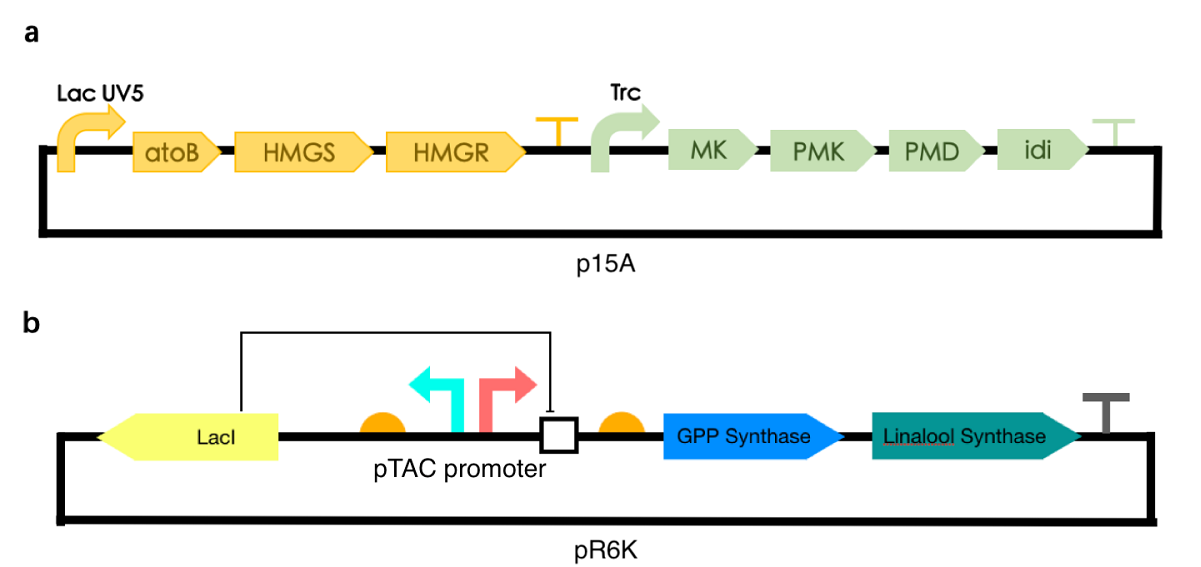
Form our experimental results, the characterization of this new part is successful. A significant linalool fragrance is produced from the E.coli which we’ve cotransformed the two plasmids into. We were able to distinguish that we did produce linalool instead of geraniol since based on our observation, the aroma of geraniol contains sweetness, whereas the aroma of standard linalool sample has a slight peppery smell, which was identifiable in our samples. The original smell of E.coli has been overcome by the fragrance of linalool and is almost neglectable. See https://2020.igem.org/Team:KEYSTONE/Engineering for more information
Reference
[1]Alonso-Gutierrez, J., Chan, R., Batth, T. S. et al (2013). Metabolic engineering of Escherichia coli for limonene and perillyl alcohol production. Metabolic Engineering, volume 19, pp.33-41. https://doi.org/10.1016/j.ymben.2013.05.004.
[2]Xun, W., Jing, W., Jiaming, C. et al (2019). Efficient biosynthesis of R-(-)-linalool through adjusting expression strategy and increasing GPP supply in Escherichia coli. https://doi.org/10.21203/rs.2.18761/v1.
