Difference between revisions of "Part:BBa K3352000"
| Line 75: | Line 75: | ||
<b> Figure 6: SDS-PAGE results show that SplintR ligase was expressed by <i>E. coli.</i> Bacterial cultures were grown overnight at 37°C, diluted to an OD600 of 0.2 and grown to 0.5, where a sample of 1mL was collected. IPTG was then added and the cultures grew for another 4 hours. After the additional 4 hours, another 1mL sample was collected. Both samples were centrifuged and the pellets were resuspended in 1x Sample Buffer. The sample with the IPTG expressed the protein more strongly, which suggests that our protein was present. SplintR was present at around 35kDa. </b> | <b> Figure 6: SDS-PAGE results show that SplintR ligase was expressed by <i>E. coli.</i> Bacterial cultures were grown overnight at 37°C, diluted to an OD600 of 0.2 and grown to 0.5, where a sample of 1mL was collected. IPTG was then added and the cultures grew for another 4 hours. After the additional 4 hours, another 1mL sample was collected. Both samples were centrifuged and the pellets were resuspended in 1x Sample Buffer. The sample with the IPTG expressed the protein more strongly, which suggests that our protein was present. SplintR was present at around 35kDa. </b> | ||
| + | |||
| + | |||
| + | <b><font size="+1.2"> Improvement of an Existing Part </font></b> | ||
| + | |||
| + | We optimized the protein coding region for Φ29 DNA polymerase by adding a N-terminal 6x histidine tag and a GS linker (BBa_K3352001). This improves the existing Φ29 DNA polymerase (BBa_K2918034) from team TUDelft 2019. To test whether our improved sequence was better, we expressed our Φ29 (BBa_K3352009), which contains (BBa_K3352001) alongside TUdelft’s (BBa_K2918034) and analyzed the expression levels by SDS-PAGE. Bacterial cultures were grown overnight at 37°C, and diluted to an OD600 of 0.2 and grown to 0.5, where a sample of 1mL was collected. We then added IPTG and grew the cultures for another 4 hours after which another 1mL sample was collected. We centrifuged All samples and resuspended the pellets in 1x Sample Buffer. In our protein gel, we see that our construct is able to better express Φ29, especially after inducing it with IPTG, relative to the pre-existing part | ||
| + | |||
| + | https://2020.igem.org/wiki/images/d/dd/T--TAS_Taipei--Registry_12.png | ||
| + | |||
| + | <b>Figure 8: Comparison between part (BBa_K2918034) and part (BBa_K3352009) showing our improved Φ29 construct. We can clearly see a band that is around 68kDa in the post-induced pET T7 promoter and Φ29 construct (red) that is not present in the post-induced TUDelft 2019 Φ29 construct (blue), which suggests that our construct can better express Φ29 DNA polymerase.</b> | ||
Revision as of 08:42, 24 October 2020
SplintR Ligase with His-Tag and GS Linker Sequence
SplintR ligase catalyzes the ligation of adjacent single-stranded DNA splinted by complementary RNA strands [4]. SplintR ligase has been previously shown to be capable of differentiating these ligation junctions to SNP levels and ligate padlock probes [4].
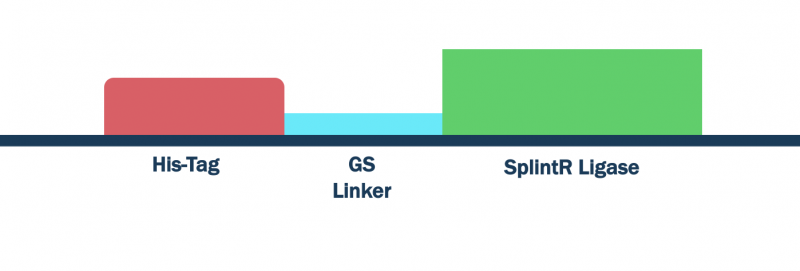
Figure 1: SplintR ligase with His-Tag and GS linker
Construct Design
We optimized the DNA sequence for expression in E. coli and removed the PstI cutting site. We attached a 6x histidine tag (6x His-Tag) upstream of the SplintR ligase sequence for purification purposes followed by a glycine-serine linker (GS linker) to form our ORF (BBa_K3352001). This served as our open reading frame (ORF; BBa_K3352000).
Results

Figure 2: Characterization of parts BBa_K3352004, BBa_K3352005, BBa_K3352006 and BBa_K3352007, which shows the Φ29 and SplintR plasmids. All four constructs were ordered from Twist or IDT, conformed to a biobrick assembly standard 10, and digested with Ecor1 and PstI. Parts BBa_K3352004 and BBa_K3352005 were ordered from IDT and had a kanamycin backbone (pUCIDT KAN) which had a size of 2.7kB. BBa_K3352007 was also ordered from IDT, however, it contained an ampicillin backbone (pUCIDT AMP) which was also around 2.7kB. BBa_K3352006 was obtained from Twist Bioscience and was cloned into the ampicillin backbone (pSB1A3).
Characterization
Strong Promoter and Strong RBS
We flanked with an upstream strong promoter and strong ribosome binding site (RBS) combination (BBa_K880005) and downstream double terminator (BBa_B0015). This entire composite part was gene synthesized by IDT.
Protein Expression and Purification
We transformed our designed plasmids into DH5⍺ E. coli cells. We grew overnight cultures, diluted those cultures, and then grew the cells to log phase. We lysed cells with xTractor Lysis Buffer (Takara Bio) and purified our His-tagged proteins using Ni sepharose affinity chromatography. In order to check if our proteins were correct, we used SDS-PAGE [6]
Based on our results, our SplintR ligase construct that used a strong promoter and strong RBS combination (BBa_K3352004 and BBa_K3352005) did not express an appreciable amount of protein (Figure 3).

Figure 3: SDS-PAGE results show protein content at different steps of protein purification. A band around 35 kDa in the cell lysate (blue) and the eluate (red), matches our expected HIS-tagged Phi29. However, many other proteins were present in the eluate, and in the flowthrough lane (yellow). There was also a similar band when there is not supposed to be one. This prompted us to redesign our constructs.
Improved Design
T7 Promoter and Strong RBS
Seeing that purified SplintR ligase is fundamental to the development of our diagnostic test, we attempted to resolve the issue of low protein expression by replacing the strong promoter in our constructs with a T7 promoter and expressing our protein in BL21(DE3) E. coli. BL21(DE3) strains contain the chromosomal gene T7 RNA polymerase, which is regulated by a lac promoter [2]. T7 RNA polymerase has been found to be highly selective and efficient in transcribing only the T7 promoter [1, 2]. Resulting in almost a five-fold faster elongation rate than E. coli RNA polymerase, T7 would be a much stronger promoter of choice. Thus, by using IPTG during protein expression to activate the lac promoter, and thus the T7 RNA polymerase, of our BL21(DE3) E. coli culture, we would effectively significantly increase the production of our enzymes positioned downstream of our T7 promoter [2,5]. We obtained the sequence of the T7 promoter (BBa_J65997) and the strong RBS (BBa_B0034) from the Parts Registry and used it to replace the strong combination on our SplintR ligase construct. This part was synthesized by Twist Biosciences and IDT.
Protein Expression and Purification
We transformed our newly designed plasmids into BL21(DE3) E. coli cells. We grew overnight cultures, diluted those cultures, then grew the cells to OD600 0.5. We then induced expression with 0.1 M IPTG and allowed cultures to grow an additional 2 hours. We harvested cells and then lysed them with xTractor Lysis Buffer [6]. We purified our His-tagged proteins using Ni sepharose affinity chromatography. In order to check if our proteins were correct, we used SDS-PAGE. Our results show SplintR ligase migrating at the expected sizes of 35.7 kDa.

Figure 4: Our SDS-PAGE results show that E. coli is able to produce SplintR ligase. Bacterial cultures were grown overnight at 37°C, lysed, and prepped for SDS-PAGE. The expected size is listed on the side.
pET3a T7 Promoter
We also aimed to improve this construct by using pET3a vectors with appropriate BioBrick prefixes and suffixes that fulfill the assembly standard. The pET vector includes the T7 promoter, which promotes high level transcription. By utilizing both a T7 promoter, T7 terminator, and an extended UTR sequence around the RBS and before the terminator, we would maximize the protein expression for our enzymes. These composite parts were synthesized by GenScript.
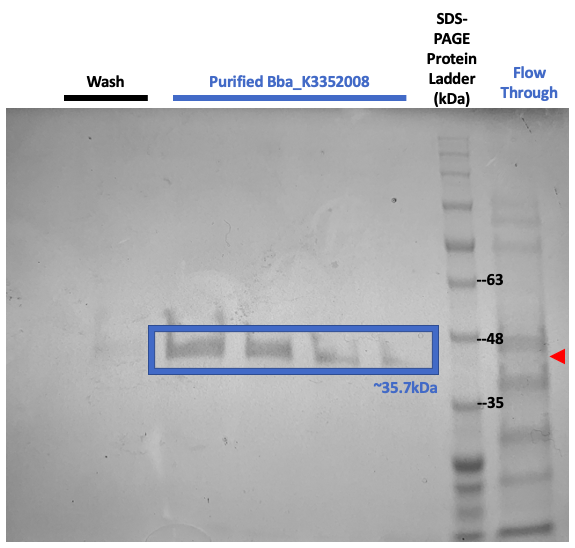
Figure 5: SDS-PAGE results show protein content at different steps of protein purification. A band around 35kDa was not present in the flow through lane (red) or the wash buffer lanes, which corresponds with our expected His-tagged SplintR.
Protein Expression and Purification

Figure 6: SDS-PAGE results show that SplintR ligase was expressed by E. coli. Bacterial cultures were grown overnight at 37°C, diluted to an OD600 of 0.2 and grown to 0.5, where a sample of 1mL was collected. IPTG was then added and the cultures grew for another 4 hours. After the additional 4 hours, another 1mL sample was collected. Both samples were centrifuged and the pellets were resuspended in 1x Sample Buffer. The sample with the IPTG expressed the protein more strongly, which suggests that our protein was present. SplintR was present at around 35kDa.
Improvement of an Existing Part
We optimized the protein coding region for Φ29 DNA polymerase by adding a N-terminal 6x histidine tag and a GS linker (BBa_K3352001). This improves the existing Φ29 DNA polymerase (BBa_K2918034) from team TUDelft 2019. To test whether our improved sequence was better, we expressed our Φ29 (BBa_K3352009), which contains (BBa_K3352001) alongside TUdelft’s (BBa_K2918034) and analyzed the expression levels by SDS-PAGE. Bacterial cultures were grown overnight at 37°C, and diluted to an OD600 of 0.2 and grown to 0.5, where a sample of 1mL was collected. We then added IPTG and grew the cultures for another 4 hours after which another 1mL sample was collected. We centrifuged All samples and resuspended the pellets in 1x Sample Buffer. In our protein gel, we see that our construct is able to better express Φ29, especially after inducing it with IPTG, relative to the pre-existing part

Figure 8: Comparison between part (BBa_K2918034) and part (BBa_K3352009) showing our improved Φ29 construct. We can clearly see a band that is around 68kDa in the post-induced pET T7 promoter and Φ29 construct (red) that is not present in the post-induced TUDelft 2019 Φ29 construct (blue), which suggests that our construct can better express Φ29 DNA polymerase.
References
1. Arnaud-Barbe, N. (1998). Transcription of RNA templates by T7 RNA polymerase. Nucleic Acids Research, 26(15), 3550–3554. https://doi.org/10.1093/nar/26.15.3550
2. Biolabs, N. E. (n.d.-a). E. coli Expression Strains | NEB. Retrieved October 22, 2020, from https://international.neb.com/products/competent-cells/e-coli-expression-strains/e-coli-expression-strains
3. Biolabs, N. E. (n.d.-b). Phi29 DNA Polymerase | NEB. Retrieved October 20, 2020, from https://international.neb.com/products/m0269-phi29-dna-polymerase
4. Biolabs, N. E. (n.d.-c). SplintR® Ligase | NEB. Retrieved October 20, 2020, from https://international.neb.com/products/m0375-splintr-ligase
5. T7 Promoter System Vectors for Highest Expression Levels in Bacteria. (n.d.). Sigma-Aldrich. Retrieved October 22, 2020, from https://www.sigmaaldrich.com/life-science/molecular-biology/cloning-and-expression/vector-systems/t7-promoter-system.html
6. XTractorTM Buffer & xTractor Buffer Kit User Manual. (n.d.). 10.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 790
- 1000COMPATIBLE WITH RFC[1000]
