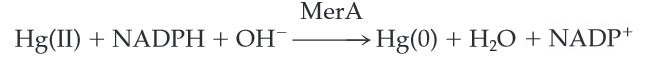Difference between revisions of "Part:BBa K1420001"
(→Overview) |
(→MIT MAHE 2020) |
||
| (85 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1420001 short</partinfo> | <partinfo>BBa_K1420001 short</partinfo> | ||
| + | |||
| + | == Summary == | ||
| + | <p>'''•''' MerA, a ~125kDa homodimeric mercuric ion reductase, is encoded by the ''merA'' gene in the ''mer'' operon.</p> | ||
| + | <p>'''•''' MerA reduces Hg(II) to Hg(0) as a means of detoxification.</p> | ||
| + | <p>'''• '''MerA and MerB (organomercury lyase) are essential to inorganic and organic mercury resistance.</p> | ||
| + | <p>'''•''' Zone of inhibition assays showed that ''E. coli'' harboring the mer operon (''E. coli'' pBBRBB::''mer'') were more resistant to mercuric ions (smallest zone of inhibition) than the vector control (''E. coli'' pBBRBB::''gfp''). Also, ''E. coli'' with the mer operon containing a deletion in ''merA'' were more sensitive to mercuric ions (larger zone of inhibition) than ''E. coli'' with the entire mer operon and the vector control. This illustrates that when other proteins encoded by the mer operon, MerT and MerP, transport mercury ions into the cell, MerA plays an important role detoxifying Hg(II) to Hg(0). | ||
| + | </p> | ||
== Overview == | == Overview == | ||
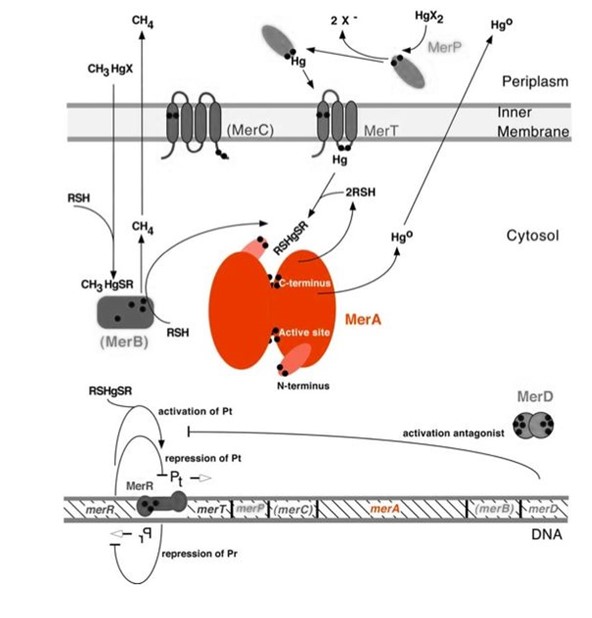
| − | <p>MerA, a | + | <p>Mercury resistance gene ''merA'' (1686 bp) encodes MerA, a mercuric ion reductase (cytosolic, flavin disulfide oxidoreductase, ~120kDa), that is essential to bacterial mercury resistance. MerA and the gene ''merA'' are highlighted in orange in ''Figure 1''. The lower part of ''Figure 1'' shows the arrangement of ''mer'' genes in the operon. ''merA'' is located downstream of ''merT'' and ''merP'', the genes that encode mercury transport proteins. As shown in ''Figure 2'', the mechanisms essential for mercury resistance are carried out by MerB and MerA. MerB transforms methylmercury to less toxic, non-biomagnifying ionic mercury Hg(II), and MerA reduces Hg(II) to the least toxic metallic mercury, Hg(0).</p> |
| + | |||
| + | [[File:MerA.jpg|center|480px|x]] | ||
<p></p> | <p></p> | ||
| − | Figure 1 shows the interactions of MerA | + | ''Figure 1.'' Model of mercury resistance operon. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. This figure shows the interactions of MerA with mercury compounds and other gene products of ''mer'' operon. (This figure and legend are adapted from "Bacterial mercury resistance from atoms to ecosystems".<sup>1</sup>) |
| − | + | ||
| + | <p>Nature evolves different mercury detoxification strategies. MerA and MerB confer mercury resistance by coupling reduction of thiol-avid organic mercury species to relatively inert, uncharged, monoatomic mercury. Since organic mercuric species have high affinity to thiol groups and commonly form strong but reversible bonds, cytosolic thiol redox buffers and cysteine residues of mercury binding proteins compete with thiol groups in other proteins and therefore prevent cationic mercury from inhibiting cytosolic machineries. Once the organomercury is converted to ionic mercury by MerB, MerA reduces ionic mercury to volatile, elemental mercury which diffuses through cell membrane without any active transport systems. While MerA and MerB are the key mercury detoxification enzymes, they orchestrate with mercury transport proteins, MerT and MerP, to confer bacterial mercury resistance.</p> | ||
<p></p> | <p></p> | ||
| − | |||
| − | == Function == | + | == Molecular Function == |
| − | MerA catalyzes the reduction of mercuric ion to the relative inert, | + | MerA catalyzes the reduction of mercuric ion released from protonlysis of organomercury (catalyzed by MerB) to the relative inert, elemental mercury in a NADPH dependent reaction (''Scheme 1''). |
| + | <p></p> | ||
| + | ''Scheme 1.'' MerA Catalyzed Reaction | ||
<p></p> | <p></p> | ||
| + | [[File:merAReactionmechanism.jpg|450px]] | ||
<p></p> | <p></p> | ||
| + | |||
<p></p> | <p></p> | ||
| − | < | + | |
| + | == Structure and Mechanism == | ||
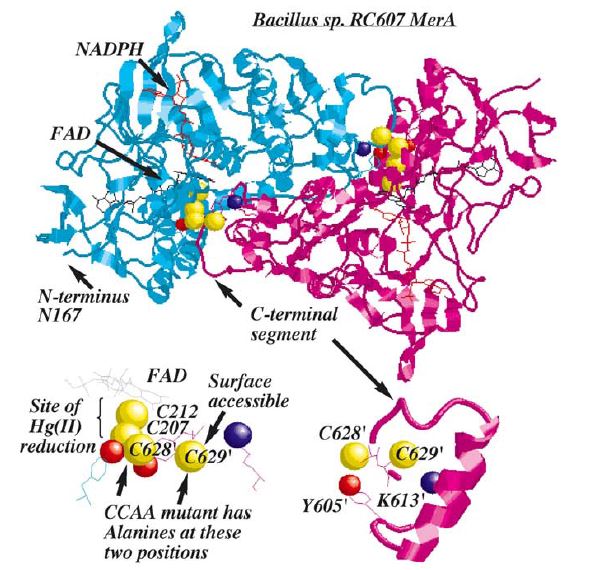
| + | ''Figure 2.'' MerA is a 125kDA homodimer. The catalytic site region of MerA is homologous to pyridine nucleotide disulfide oxireductases in which the homologous active site region formed by two cysteines. Hg(II) forms a complex with these two inner cysteines. In addition to the active site, two unique regions, each with a pair of cysteines, although not essential to Hg(II) reduction, are required for transporting Hg(II) from solution to catalytic core. These two unique regions are (1) a short, mobile C-terminal extension and (2)a long N-terminal extension (95aa). Based on crystal structure and mutagenesis studies, C-terminal extension of a monomer is in close proximity to two inner cysteines from the other monomer, providing the acquisition of Hg(II) from the solution to active site. Although the pair of cysteines in C-terminal extension is not essential in Hg(II) reduction, they provide an essential ligand exchange pathway that removes thiolate before Hg(II) binds to active site. This prevents accumulation of negative charge build up at active site. | ||
| + | N-terminal extension, which also does not form a complex in the catalytic core, serves as a facilitator for Hg(II) aquisition and delivery to the core. Research has shown 70aa of this domain were homologous to MerP, the periplasmic mercuric binding protein, which adopt ferredoxin-like beta-alpha-beta-beta-alpha-beta structural fold and use the cysteines to bind metal ions. It is further elucidated that N-terminal extension binds a single equivalent of Hg(II) using the two cysteines, and this Hg(II)-domain complex is an substrate for the catalytic core in the absence of thiol-containing compound<sup>1</sup>. | ||
<p></p> | <p></p> | ||
<p></p> | <p></p> | ||
| − | + | [[File:StructureMerA.jpg|400px|]] | |
| + | <p></p><p></p><p></p> | ||
<p></p> | <p></p> | ||
| − | |||
| + | == Characterization of ''merA'' == | ||
| + | To test the effect of MerA on mercury resistance, we characterized mercury resistance or sensitivity by zone of inhibition tests using the following three strains: | ||
| + | <p>'''•'''''E. coli'' K12 pBBRBB::''gfp'' - vector control</p> | ||
| + | <p>'''•'''''E. coli'' K12 pBBRBB::''mer'' - mer operon vector (''merRTPAB'')</p> | ||
| + | <p>'''•'''''E. coli'' K12 pBBRBB::''mer''Δ''merA''- mer operon with ''merA'' deleted (''merRTPB'')</p> | ||
| + | |||
| − | + | <p></p> | |
| + | <b>Zone of Inhibition: Mercury(II) Chloride Plate Test Results</b> | ||
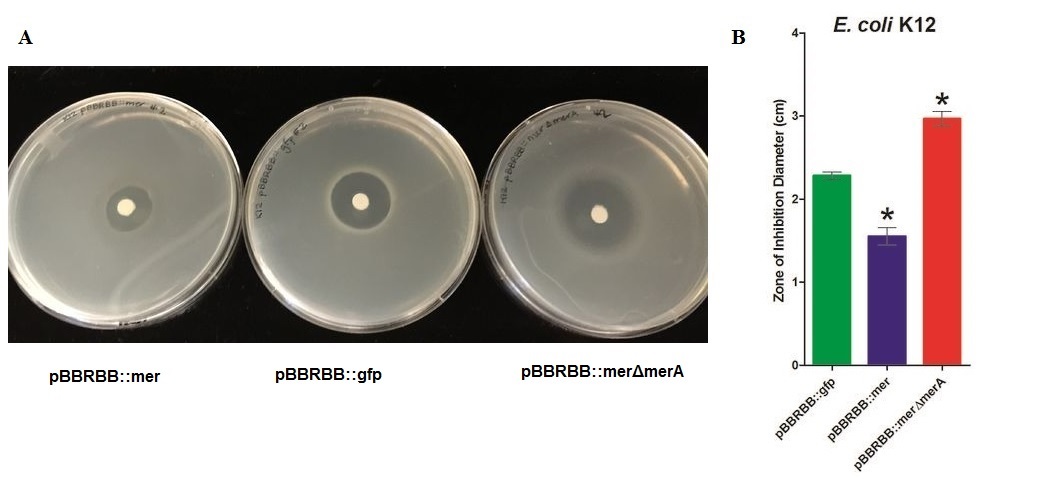
| + | [[File:2MerA ZOI K12.jpg|950px|center|alt text]] | ||
| − | + | ''Figure 3.'' Zones of Inhibition Test For Mercury Resistance. In this assay, ''Escherichia coli'' K12 expressing three different constructs was spread on agar plates to compare level of mercury resistance. Each agar plate contained a filter disk spotted with 10µL of 0.1M HgCl2 in the middle. (A) Agar Plates of K12 with each construct. Left, K12 containing pBBRBB::''mer'' (''mer'' operon); center, K12 containing pBBRBB::''gfp'' (vector control); right, K12 containing pBBRBB::''mer''Δ''merA''(''mer'' operon with ''merA'' deleted). (B) Diameter of zones of inhibition. The diameter of the zone of inhibition was measured in triplicate. Green corresponds to pBBRBB::''gfp'', blue to pBBRBB::''merRTPAB'', and red to pBBRBB::''mer''Δ''merA''. Individual constructs range from highest resistance in the following order: ''merRTPAB'' > vector control >''mer''Δ''merA''. | |
| − | + | ||
| − | + | For protocol of zone inhibition test, please click [[Mercury(II) Chloride Plate test protocol]]. | |
| − | + | ||
| − | <p></p> | + | ''E. coli'' K12 containing the plasmid with the mer operon (pBBRBB:''merRTPAB'') had a smaller zone of inhibition than ''E. coli'' with the vector control indicating that expression of the ''mer'' operon increased resistance to Hg(II) (i.e. allowed cells to grow closer to the Hg(II)-containing disc resulting in a smaller zone of inhibition). ''E. coli'' cells harboring the plasmid containing the mer operon with ''merA'' deleted had the largest zones of inhibition among all three strain tested. This result was expected since cells containing ''merRTPB'' without ''merA'' are able to transport Hg(II) more efficiently into the cell but cannot reduce Hg(II) to the less toxic Hg(0). Therefore, Hg(II) accumulates in cells lacking MerA and hence they are unable to grow near the Hg(II)-containing disc. |
| − | < | + | |
| − | + | These results demonstrate the role of ''merA'' in the context of the entire operon. For future directions, we are working to clone ''merA'' in a vector containing a different origin of replication than pBBRBB. This way we can express ''merA'' ''in trans'' alongside pBBRBB::''merΔmerA'' to ensure the part complements the deletion and tests the part more directly. Since expression level of ''merA'' is tightly regulated both by MerR and overall position in the operon as a whole, this will likely need to be done using a low copy or inducible vector. | |
| + | |||
| + | ==MIT_MAHE 2020== | ||
| + | |||
| + | '''Usage and Biology''' | ||
| + | |||
| + | Partial peptide sequences confirmed the relationship by the presence of a highly homologous active site region with the two cysteines that form the characteristic redox-active disulfide/dithiol of the family. However, unlike the other family members that use this cysteine pair for catalysis and are inhibited when Hg(II) binds to them, MerA has additional structural features to help it avoid such inhibition. | ||
| + | |||
| + | Results from studies show that the impact of the NmerA domain on the kinetics increases as the size of the thiol ligand on the Hg(II) substrate increases and as the excess concentration of the thiol ligand in the assay decreases (Ledwidge, Falkowski and Miller, unpublished results). This suggests that the NmerA domain may only become critical under conditions of depletion of the cellular thiol pool such that binding of Hg(II) to protein thiols would then become more problematic. Since the in vivo function of the N-terminal di-Ala mutant was tested in otherwise unstressed E. coli, the normal intracellular thiol concentrations of GSH (ca. 6 mM) may have eliminated any benefit the NmerA domain could provide. This proposal is currently under investigation using GSH depleted cells (Summers and Miller, unpublished results). | ||
| + | |||
| + | The correlation of double repeat sequences with low intracellular thiol concentration strongly suggests that the NmerA domains should be more critical for resistance in those organisms since they must serve both as the intracellular buffer to prevent inhibitory binding to other proteins in the cell and as the specific delivery agent to the catalytic core. This idea further suggests that in cells with higher concentrations of thiols, the single domain would not normally be needed as the buffering agent, but might become important in this role if the cells are subjected to additional oxidative or other stresses that deplete the intracellular thiol concentration. | ||
| + | |||
| + | ==References== | ||
| + | <p>1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." ''FEMS Microbiology Reviews'' <b>27</b>: 355-384.</p> | ||
| + | <p>2. V. B. Mathema et al (2011). "Bacterial ''mer'' operon-mediated detoxification of mercurial compounds: a short review". ''Archives of Mictobiology'' <b>193</b>: 837-844.</p> | ||
| + | <p>3. Annette K. Møller, Tamar Barkay, Martin A. Hansen, Anders Norman, Lars H. Hansen, Søren J. Sørensen, Eric S. Boyd, Niels Kroer, Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine, FEMS Microbiology Ecology, Volume 87, Issue 1, January 2014, Pages 52–63, https://doi.org/10.1111/1574-6941.12189 </p> | ||
| − | + | <p> 4. Moore, M. J., Distefano, M. D., Zydowsky, L. D., Cummings, R. T., & Walsh, C. T. (1990). Organomercurial lyase and mercuric ion reductase: nature’s mercury detoxification catalysts. Accounts of Chemical Research, 23(9), 301–308. https://doi.org/10.1021/ar00177a006 </p> | |
| − | + | ||
| + | <p> 5. Benison, G. C., Di Lello, P., Shokes, J. E., Cosper, N. J., Scott, R. A., Legault, P., & Omichinski, J. G. (2004). A stable mercury-containing complex of the organomercurial lyase MerB: catalysis, product release, and direct transfer to MerA. Biochemistry, 43(26), 8333–8345. https://doi.org/10.1021/bi049662h </p> | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 18:56, 23 October 2020
merA, mercuric reductase from Serratia marcescens
Summary
• MerA, a ~125kDa homodimeric mercuric ion reductase, is encoded by the merA gene in the mer operon.
• MerA reduces Hg(II) to Hg(0) as a means of detoxification.
• MerA and MerB (organomercury lyase) are essential to inorganic and organic mercury resistance.
• Zone of inhibition assays showed that E. coli harboring the mer operon (E. coli pBBRBB::mer) were more resistant to mercuric ions (smallest zone of inhibition) than the vector control (E. coli pBBRBB::gfp). Also, E. coli with the mer operon containing a deletion in merA were more sensitive to mercuric ions (larger zone of inhibition) than E. coli with the entire mer operon and the vector control. This illustrates that when other proteins encoded by the mer operon, MerT and MerP, transport mercury ions into the cell, MerA plays an important role detoxifying Hg(II) to Hg(0).
Overview
Mercury resistance gene merA (1686 bp) encodes MerA, a mercuric ion reductase (cytosolic, flavin disulfide oxidoreductase, ~120kDa), that is essential to bacterial mercury resistance. MerA and the gene merA are highlighted in orange in Figure 1. The lower part of Figure 1 shows the arrangement of mer genes in the operon. merA is located downstream of merT and merP, the genes that encode mercury transport proteins. As shown in Figure 2, the mechanisms essential for mercury resistance are carried out by MerB and MerA. MerB transforms methylmercury to less toxic, non-biomagnifying ionic mercury Hg(II), and MerA reduces Hg(II) to the least toxic metallic mercury, Hg(0).
Figure 1. Model of mercury resistance operon. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. This figure shows the interactions of MerA with mercury compounds and other gene products of mer operon. (This figure and legend are adapted from "Bacterial mercury resistance from atoms to ecosystems".1)
Nature evolves different mercury detoxification strategies. MerA and MerB confer mercury resistance by coupling reduction of thiol-avid organic mercury species to relatively inert, uncharged, monoatomic mercury. Since organic mercuric species have high affinity to thiol groups and commonly form strong but reversible bonds, cytosolic thiol redox buffers and cysteine residues of mercury binding proteins compete with thiol groups in other proteins and therefore prevent cationic mercury from inhibiting cytosolic machineries. Once the organomercury is converted to ionic mercury by MerB, MerA reduces ionic mercury to volatile, elemental mercury which diffuses through cell membrane without any active transport systems. While MerA and MerB are the key mercury detoxification enzymes, they orchestrate with mercury transport proteins, MerT and MerP, to confer bacterial mercury resistance.
Molecular Function
MerA catalyzes the reduction of mercuric ion released from protonlysis of organomercury (catalyzed by MerB) to the relative inert, elemental mercury in a NADPH dependent reaction (Scheme 1).
Scheme 1. MerA Catalyzed Reaction
Structure and Mechanism
Figure 2. MerA is a 125kDA homodimer. The catalytic site region of MerA is homologous to pyridine nucleotide disulfide oxireductases in which the homologous active site region formed by two cysteines. Hg(II) forms a complex with these two inner cysteines. In addition to the active site, two unique regions, each with a pair of cysteines, although not essential to Hg(II) reduction, are required for transporting Hg(II) from solution to catalytic core. These two unique regions are (1) a short, mobile C-terminal extension and (2)a long N-terminal extension (95aa). Based on crystal structure and mutagenesis studies, C-terminal extension of a monomer is in close proximity to two inner cysteines from the other monomer, providing the acquisition of Hg(II) from the solution to active site. Although the pair of cysteines in C-terminal extension is not essential in Hg(II) reduction, they provide an essential ligand exchange pathway that removes thiolate before Hg(II) binds to active site. This prevents accumulation of negative charge build up at active site. N-terminal extension, which also does not form a complex in the catalytic core, serves as a facilitator for Hg(II) aquisition and delivery to the core. Research has shown 70aa of this domain were homologous to MerP, the periplasmic mercuric binding protein, which adopt ferredoxin-like beta-alpha-beta-beta-alpha-beta structural fold and use the cysteines to bind metal ions. It is further elucidated that N-terminal extension binds a single equivalent of Hg(II) using the two cysteines, and this Hg(II)-domain complex is an substrate for the catalytic core in the absence of thiol-containing compound1.
Characterization of merA
To test the effect of MerA on mercury resistance, we characterized mercury resistance or sensitivity by zone of inhibition tests using the following three strains:
•E. coli K12 pBBRBB::gfp - vector control
•E. coli K12 pBBRBB::mer - mer operon vector (merRTPAB)
•E. coli K12 pBBRBB::merΔmerA- mer operon with merA deleted (merRTPB)
Zone of Inhibition: Mercury(II) Chloride Plate Test Results
Figure 3. Zones of Inhibition Test For Mercury Resistance. In this assay, Escherichia coli K12 expressing three different constructs was spread on agar plates to compare level of mercury resistance. Each agar plate contained a filter disk spotted with 10µL of 0.1M HgCl2 in the middle. (A) Agar Plates of K12 with each construct. Left, K12 containing pBBRBB::mer (mer operon); center, K12 containing pBBRBB::gfp (vector control); right, K12 containing pBBRBB::merΔmerA(mer operon with merA deleted). (B) Diameter of zones of inhibition. The diameter of the zone of inhibition was measured in triplicate. Green corresponds to pBBRBB::gfp, blue to pBBRBB::merRTPAB, and red to pBBRBB::merΔmerA. Individual constructs range from highest resistance in the following order: merRTPAB > vector control >merΔmerA.
For protocol of zone inhibition test, please click Mercury(II) Chloride Plate test protocol.
E. coli K12 containing the plasmid with the mer operon (pBBRBB:merRTPAB) had a smaller zone of inhibition than E. coli with the vector control indicating that expression of the mer operon increased resistance to Hg(II) (i.e. allowed cells to grow closer to the Hg(II)-containing disc resulting in a smaller zone of inhibition). E. coli cells harboring the plasmid containing the mer operon with merA deleted had the largest zones of inhibition among all three strain tested. This result was expected since cells containing merRTPB without merA are able to transport Hg(II) more efficiently into the cell but cannot reduce Hg(II) to the less toxic Hg(0). Therefore, Hg(II) accumulates in cells lacking MerA and hence they are unable to grow near the Hg(II)-containing disc.
These results demonstrate the role of merA in the context of the entire operon. For future directions, we are working to clone merA in a vector containing a different origin of replication than pBBRBB. This way we can express merA in trans alongside pBBRBB::merΔmerA to ensure the part complements the deletion and tests the part more directly. Since expression level of merA is tightly regulated both by MerR and overall position in the operon as a whole, this will likely need to be done using a low copy or inducible vector.
MIT_MAHE 2020
Usage and Biology
Partial peptide sequences confirmed the relationship by the presence of a highly homologous active site region with the two cysteines that form the characteristic redox-active disulfide/dithiol of the family. However, unlike the other family members that use this cysteine pair for catalysis and are inhibited when Hg(II) binds to them, MerA has additional structural features to help it avoid such inhibition.
Results from studies show that the impact of the NmerA domain on the kinetics increases as the size of the thiol ligand on the Hg(II) substrate increases and as the excess concentration of the thiol ligand in the assay decreases (Ledwidge, Falkowski and Miller, unpublished results). This suggests that the NmerA domain may only become critical under conditions of depletion of the cellular thiol pool such that binding of Hg(II) to protein thiols would then become more problematic. Since the in vivo function of the N-terminal di-Ala mutant was tested in otherwise unstressed E. coli, the normal intracellular thiol concentrations of GSH (ca. 6 mM) may have eliminated any benefit the NmerA domain could provide. This proposal is currently under investigation using GSH depleted cells (Summers and Miller, unpublished results).
The correlation of double repeat sequences with low intracellular thiol concentration strongly suggests that the NmerA domains should be more critical for resistance in those organisms since they must serve both as the intracellular buffer to prevent inhibitory binding to other proteins in the cell and as the specific delivery agent to the catalytic core. This idea further suggests that in cells with higher concentrations of thiols, the single domain would not normally be needed as the buffering agent, but might become important in this role if the cells are subjected to additional oxidative or other stresses that deplete the intracellular thiol concentration.
References
1. T. Barkay et al (2003). "Bacterial mercury resistance from atoms to ecosystems." FEMS Microbiology Reviews 27: 355-384.
2. V. B. Mathema et al (2011). "Bacterial mer operon-mediated detoxification of mercurial compounds: a short review". Archives of Mictobiology 193: 837-844.
3. Annette K. Møller, Tamar Barkay, Martin A. Hansen, Anders Norman, Lars H. Hansen, Søren J. Sørensen, Eric S. Boyd, Niels Kroer, Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine, FEMS Microbiology Ecology, Volume 87, Issue 1, January 2014, Pages 52–63, https://doi.org/10.1111/1574-6941.12189
4. Moore, M. J., Distefano, M. D., Zydowsky, L. D., Cummings, R. T., & Walsh, C. T. (1990). Organomercurial lyase and mercuric ion reductase: nature’s mercury detoxification catalysts. Accounts of Chemical Research, 23(9), 301–308. https://doi.org/10.1021/ar00177a006
5. Benison, G. C., Di Lello, P., Shokes, J. E., Cosper, N. J., Scott, R. A., Legault, P., & Omichinski, J. G. (2004). A stable mercury-containing complex of the organomercurial lyase MerB: catalysis, product release, and direct transfer to MerA. Biochemistry, 43(26), 8333–8345. https://doi.org/10.1021/bi049662h
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1201
Illegal NgoMIV site found at 1249
Illegal NgoMIV site found at 1311
Illegal NgoMIV site found at 1522 - 1000COMPATIBLE WITH RFC[1000]