Difference between revisions of "Part:BBa K3453143"
| Line 3: | Line 3: | ||
<partinfo>BBa_K3453143 short</partinfo> | <partinfo>BBa_K3453143 short</partinfo> | ||
| − | + | This part is a transcription unit of the Rosewood DmRbcL Trigger 1.3 ([[Part:BBa_K3453043|BBa_K3453043]]) for sequence-based detection of rosewood. The transcription is controlled by the T7 promoter ([[Part:BBa_K2150031|BBa_K2150031]]) and the strong SBa_000587 synthetic terminator ([[Part:BBa_K3453000|BBa_K3453000]]). | |
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
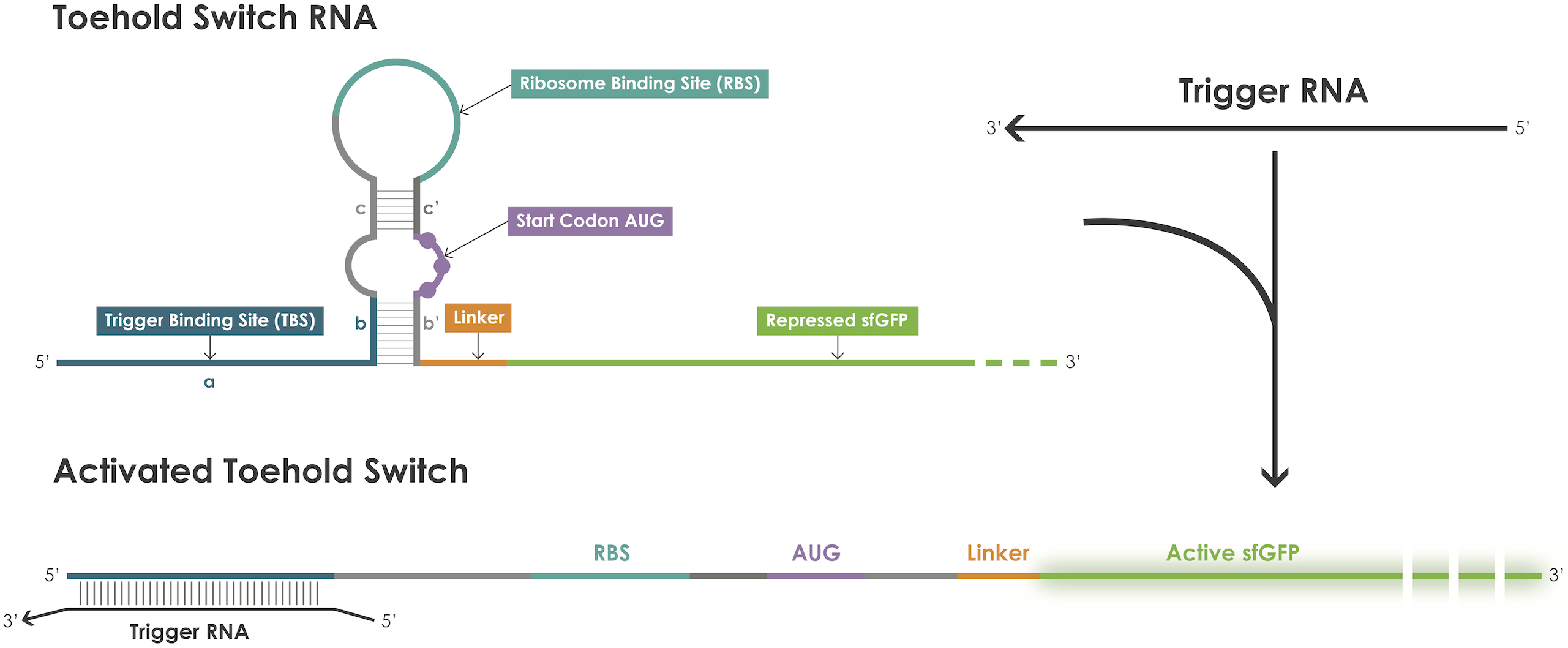
| + | A toehold switch is an RNA–based device containing a ribosome binding site (RBS) and an ATG start codon embedded in a hairpin structure that blocks translation initiation [1]. The hairpin can be unfolded upon binding of a trigger RNA therefore exposing the RBS and the ATG start codon permitting translation of the reporter protein (Figure 1). | ||
| + | |||
| + | [[File: T--Evry_Paris-Saclay--toehold-switch.png|600px]] | ||
| + | |||
| + | Figure 1. Toehold switches principle. | ||
| + | |||
| + | This part is a transcription unit of [[Part:BBa_K3453043|BBa_K3453043]] which is a trigger for the Rosewood DmRbcL Toehold Switch 1.3 ([[Part:BBa_K3453033|BBa_K3453033]]) and also contains the switch binding site for the Rosewood DmRbcL Toehold Switch 2.1 ([[Part:BBa_K3453034|BBa_K3453034]]). | ||
| + | |||
| + | This part proved to be functional: the transcribed rosewood RNA was able to bind to the corresponding toehold switches and a readable output was generated. | ||
| + | |||
| + | Full results are available on the [[Part:BBa_K3453133|BBa_K3453133]] and [[Part:BBa_K3453134|BBa_K3453134]] pages in the registry. | ||
| + | |||
| + | ===References=== | ||
| + | [1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939. | ||
<!-- --> | <!-- --> | ||
Revision as of 18:22, 18 October 2020
Rosewood DmRbcL Trigger 1.3 transcription unit
This part is a transcription unit of the Rosewood DmRbcL Trigger 1.3 (BBa_K3453043) for sequence-based detection of rosewood. The transcription is controlled by the T7 promoter (BBa_K2150031) and the strong SBa_000587 synthetic terminator (BBa_K3453000).
Usage and Biology
A toehold switch is an RNA–based device containing a ribosome binding site (RBS) and an ATG start codon embedded in a hairpin structure that blocks translation initiation [1]. The hairpin can be unfolded upon binding of a trigger RNA therefore exposing the RBS and the ATG start codon permitting translation of the reporter protein (Figure 1).
Figure 1. Toehold switches principle.
This part is a transcription unit of BBa_K3453043 which is a trigger for the Rosewood DmRbcL Toehold Switch 1.3 (BBa_K3453033) and also contains the switch binding site for the Rosewood DmRbcL Toehold Switch 2.1 (BBa_K3453034).
This part proved to be functional: the transcribed rosewood RNA was able to bind to the corresponding toehold switches and a readable output was generated.
Full results are available on the BBa_K3453133 and BBa_K3453134 pages in the registry.
References
[1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]